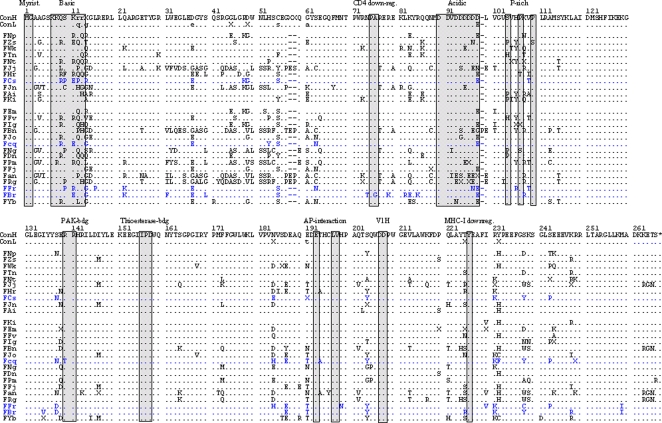
Figure 1. SIVsmm Nef sequences do not contain obvious defects.
The SIVsmm consensus Nef amino acid sequences from the CD4high (ConH) and CD4low (ConL) animals are shown in the upper panels. Nef sequences derived from the four experimentally infected SMs are shown in blue. The order of the SIVsmm Nef alleles corresponds to that of the CD4+ T cell counts in the respective SMs; i.e. the Nef sequence from animal FNp with the highest numbers shown at the top and from FYb that showest the most severe CD4+ T cell depletion at the bottom. Some conserved sequence elements in Nef, including the N-terminal myristoylation signal, N-proximal tyrosines, PA residues critical for CD4 down-modulation by SIVmac Nef, the basic, acidic and proline-rich regions, a diarginine motif, a C-proximal adaptor-protein (AP) interaction site, a diacidic putative V1H binding site and a Y residue involed in MHC-I downregulation are indicated. Dots specify identity with the consensus Nef sequence and dashes gaps introduced to optimize the alignment. PCR-fragments amplified from SM plasma were sequenced directly and “X” indicates nucleotide ambiguity in the corresponding codons.