Abstract
Nuclear tRNA aminoacylation was proposed to provide a proofreading step in Xenopus oocytes, ensuring nuclear export of functional tRNAs [Lund, E. & Dahlberg, J. E. (1998) Science 282, 2082–2085]. Herein, it is documented that tRNA aminoacylation also occurs in yeast nuclei and is important for tRNA export. We propose that tRNA aminoacylation functions in one of at least two parallel paths of tRNA export in yeast. Alteration of one aminoacyl-tRNA synthetase affects export of only cognate tRNA, whereas alterations of two other aminoacyl-tRNA synthetases affect export of both cognate and noncognate tRNAs. Saturation of tRNA export pathway is a possible explanation of this phenomenon.
Keywords: yeast, exportin-t
RNAs must move from their site of transcription in the nucleus to the cytosol where they participate in protein synthesis. They are exported via a saturable process through large structures, nuclear pores, connecting the nuclear and cytosolic compartments (ref. 1; for review, see ref. 2). Multiple RNA-class-specific export pathways are now known to be responsible for the export of various types of RNAs (ref. 3; for review, see ref. 4).
Our studies concern the export of tRNAs to the cytosol. tRNAs are transcribed as precursor molecules and undergo many RNA-processing steps while in the nucleus. These include removal of excess 5′- and 3′-located transcribed sequences, removal of transcribed introns, numerous nucleoside modifications, and addition of CCA to the 3′ terminus (for review, see ref. 5). The tRNAs are exported to the cytosol after processing. Some components important to their nuclear export have been identified. The GTPase, Ran, and its regulators function in export and import of most proteins and RNAs, including tRNAs (for review, see ref. 6; ref. 7). Another factor is exportin-t, a member of the importin-β family (8), which binds tRNA and is thought to function in tRNA nuclear export (9, 10). Yeast Los1p is the exportin-t homologue (7, 8, 11). However, as LOS1 is not essential (12), other paths of tRNA export are likely to exist, at least in Saccharomyces cerevisiae.
Studies conducted with Xenopus oocytes led to the proposal that a nucleus-located proofreading system also functions in tRNA export, ensuring that predominately functional tRNAs leave the nuclear interior (13). According to Lund and Dahlberg (13), proofreading is achieved by the action of nucleus-located aminoacyl-tRNA synthetases. Others have suggested that exportin-t provides the tRNA proofreading function, because in vitro binding studies show that exportin-t is able to interact with uncharged tRNAs and that these interactions are best with the fully modified, end-matured tRNA (14, 15). In addition, a mutant tRNA, unable to be charged but able to interact with exportin-t, is exported to the cytosol, albeit less well than nonmutant tRNAs (14). Clearly, additional studies are required to achieve an understanding of tRNA nuclear export.
Taking advantage of the elegant genetics that is available for yeast, we provide several lines of genetic and in vivo biochemical evidence to show that in yeast tRNAs are aminoacylated while in the nucleus and that aminoacylation is important for their nuclear export. We further show that mutations of some aminoacyl-tRNA synthetases can inhibit nuclear export of noncognate tRNAs.
Materials and Methods
Strains and Media.
The following yeast strains were used: W303 (MATα ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112); SWY27 (MATα ade2-1 ura3-1 his3-11, 15 trp1-1 leu2-3, 112 nup116-5∷HIS3; ref. 16); A364A (MATa ade1 ade2 ura1 his7 lys2 tyr1 gal1); ts341 (MATa ade1 ade2 ura1 his7 lys2 tyr1 gal1 ils1-1; ref. 17; ts indicates temperature-sensitive); SS328 (MATα ade2-101 his3Δ200 lys2-801 ura3-52); ts19:3:4 (MATa ade1 leu2 his5 lys11 gal1 gal2 mes1-1; ref. 18); NT33-5/YCpcca-M1 (relevant genotype: ura3-52 leu2-3, 112 cca1-1 plus YCpcca-M1 encoding the catalytically active cytosolic and nucleus-located Cca1p isozymes; ref. 19); and NT33-5/YCp50 (relevant genotype ura3-52 leu2-3, 112 cca1-1 plus YCp50; this strain was derived from NT33-5/YCpcca-M1 by plasmid substitution). W303 and A364A are parents of SWY27 and ts341, respectively, whereas ts19:3:4 is closely related to A364A. SS328 is the parent of strain ts2, described below.
Cloning of TYS1.
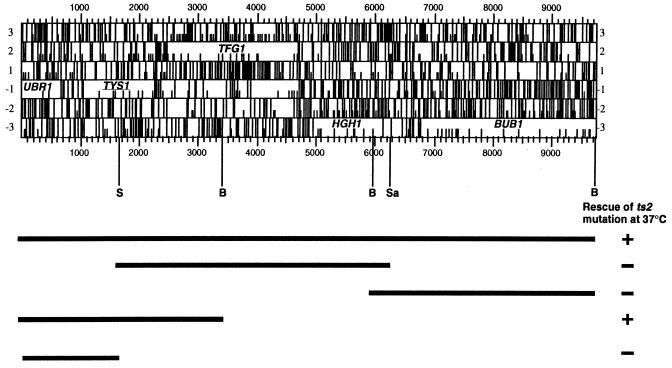
TYS1 was cloned by complementation of ts− growth defect of the mutant strain (ts2) with a genomic library constructed in YCp50 (20). One plasmid was obtained whose presence was required for growth of ts2 at 37°C. DNA sequencing of plasmid/insertion junctions determined the yeast genes encoded by the plasmid. The following oligonucleotides were used for DNA sequencing: 5′-GCCACTATCGACTACGCGAT-3′ and 5′-CCAGCAACCGCACCTGT-3′. Several ORFs—UBR1, TYS1, TFG1, HGH1, and BUB1—were contained within the insert. To identify the gene responsible for rescue of ts2 growth defects, several subclones were generated. BUB1 was cloned by BamHI digestion. A SalI–SacI fragment was generated to clone both TFG1 and HGH1 genes. Digestion of the original plasmid by BamHI generated a deletion containing TYS1 and a part of UBR1. Digestion of the original plasmid with SalI generated a plasmid containing only part of UBR1.
Oligonucleotide Probes.
We have published the sequence of the probes complementary to poly(A) RNA and tRNAIleAAU previously (7). The sequences of the probes complementary to the tRNAMet and tRNATyr were 5′-CCAGGGGAGGTTCGAACTCTCGACCTTCAGATTATGAGACTGACGCTCTTCCTACTGAGC-3′ and 5′-GCGAGTCGAACGCCCGATCTCAAGATTTACAGTCTTGCGCCTTAAACCAACTTGGCTACC-3′, respectively. All oligonucleotides used as probes for fluorescence in situ hybridization (FISH) and Northern analysis were synthesized by the Pennsylvania State University College of Medicine Macromolecular Core Facility.
FISH.
FISH was performed according to our published protocol (7). Fluorescence images were obtained by using a Nikon Microphot-FX microscope (Nikon) equipped with a SenSys charged-coupled device camera (Photometrics, Tucson, AZ). Image processing was done with qed software (Pittsburgh, PA) and adobe photoshop (Adobe Systems, Mountain View, CA). Quantitation of nuclear and cytosolic tRNA signal intensities for individual cells was performed by employing the adobe photoshop image histogram analysis algorithm. The reported percentage of nuclear tRNA in nup116Δ cells is likely to be an underestimate, because the intensity of nuclear staining in these cells is often saturating.
RNA Analysis.
Nonaminoacylated tRNAs were isolated by phenol extraction from log phase yeast cells (21). This procedure yields only small RNAs. tRNAs isolated with this method are primarily uncharged. Aminoacylated tRNAs were obtained by using a modification of a procedure previously described (22). Log phase cells were quickly chilled and collected by centrifugation at 4°C. The cells were resuspended in 0.3 M sodium acetate/10 mM sodium EDTA (pH 4.5) at 4°C, and all subsequent steps were conducted in the cold. In addition to stabilizing the aminoacyl tRNA bond, these conditions inhibit aminoacylation in the cell lysate (23). Cells were disrupted by using acid-washed glass beads. RNA was extracted by using phenol equilibrated to pH 4.5. This procedure yields rRNAs, mRNAs, as well as charged tRNAs. RNAs (20 μg) were separated by electrophoresis on a 10% polyacrylamide (pH 4.5) gel containing 8 M urea. Electroblotting and subsequent steps were performed as described (24), except that RNAs were transferred onto Hybond N+ membrane (Amersham Pharmacia).
In Vivo Labeling.
Log phase cells grown at 23°C were transferred to medium containing minimal uracil (0.1 μg/ml). Growth continued at 23°C for 90 min before the shift to 37°C. After 10 min or 60 min at 37°C, 100 μCi (1 Ci = 37 GBq) of [5,6-3H]uracil (ICN) was added. Cells were harvested 60 min later in the absence or, where indicated, presence of a 10-fold excess of nonaminoacylated yeast tRNA (Sigma). The amount of competitor tRNA added was based on tRNA being about 20% of the total RNA in yeast cells. tRNAs were isolated as described above. tRNA (2 × 105 to 4 × 105 cpm per well) was loaded. Radioactive RNAs were detected by fluorography.
Results and Discussion
Yeast Nuclear tRNAs Are Aminoacylated.
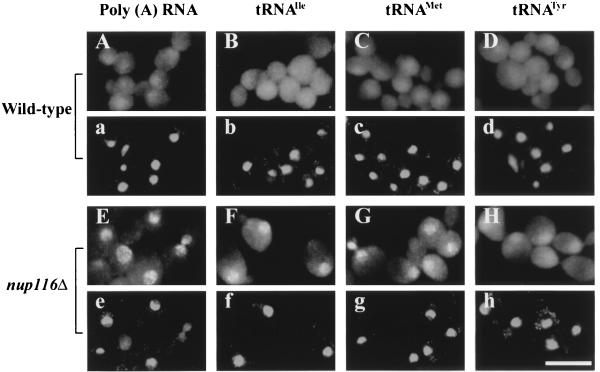
To study the role of aminoacylation in tRNA nuclear export in S. cerevisiae, we determined the subcellular location of endogenous individual tRNAs by using FISH (7). Yeast cells harboring a deletion of the gene for nucleoporin Nup116p have defective nuclear pores at elevated temperatures resulting in bidirectional inhibition of nucleus/cytosol exchange (16). Poly(A) RNA, tRNAIleAAU, tRNAMet, and tRNATyr are located primarily in the cytosol in the cells of parent strain W303, whereas they accumulate in the nucleus in nup116Δ cells (Fig. 1, compare A–D to E–H). The accumulation of nucleus-located tRNA in nup116Δ cells is significant in less than 1 h (7) and very prominent by 2 h at the nonpermissive temperature of 37°C (Fig. 1 F–H). Quantitation of the intensity of nuclear and cytosolic signals after 2 h at the nonpermissive temperature shows that an average of ≈32% of the tRNAIleAAU signal is located in the nucleus of nup116Δ cells. In contrast, under these conditions, little cellular tRNA (≈14% for tRNAIleAAU) appears to be located in nuclei of wild-type cells.
Figure 1.
Nuclear accumulation of tRNAs in nup116Δ cells. Parent W303 (A–D) and nup116Δ strain SWY27 (E–H) were grown at 23°C, and log phase cells were shifted to 37°C for 2 h. FISH was used to detect poly(A) RNA (A and E), tRNAIle (B and F), tRNAMet (C and G), and tRNATyr (D and H). a–h are 4′,6-diamidino-2-phenylindole staining of cells shown in A–H, respectively. (Bar = 10 μm.)
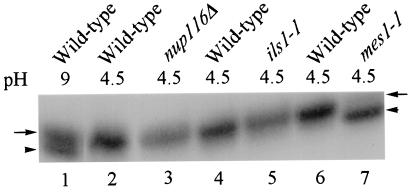
If aminoacylation is restricted to the cytosol in yeast, then greater than 30% of tRNAs isolated from nup116Δ cells at 37°C should not be aminoacylated. In contrast, if aminoacylation also can occur in the nuclear compartment, all the cellular tRNAs should be aminoacylated, including those that are confined to the nucleus. To assess the in vivo aminoacylation status of total cellular tRNAs, RNAs were isolated at 4°C and pH 4.5. These conditions stabilize the bond between the tRNA and the amino acid (22) and greatly inhibit in vitro aminoacylation in the extract (23). RNAs resolved on acidic polyacrylamide gels (22) were subjected to Northern blot analysis with a probe complementary to tRNAMet. To be certain we could distinguish the mobilities of charged and uncharged tRNAs, RNAs were also isolated from wild-type cells by using basic conditions (pH 9). This procedure generated two tRNAMet species, a slower migrating aminoacylated tRNA and a faster, nonaminoacylated tRNA, indicating partial deacylation of tRNAMet at pH 9, as expected (Fig. 2, lane 1; ref. 25). We also isolated tRNA from yeast strain ts19:3:4 (mes1-1) encoding a ts methionyl-tRNA synthetase (18). This process generates predominately the faster migrating tRNA, indicating a nearly complete block in aminoacylation with methionine (Fig. 2, lane 7). As the thermosensitivity of mes1-1 is fully reversible (greater than 90% viability after incubation at 37°C for 2 h), the lack of charged tRNAMet also indicates that there is little in vitro tRNA charging under the conditions we use.
Figure 2.
Aminoacylation of steady-state tRNAMet. tRNAs were isolated from W303 at pH 9 (lane 1) and at pH 4.5 (lane 2); SWY27, nup116Δ (lane 3); A364A (lane 4); ils1-1 (lane 5); SS328 (lane 6); and mes1-1 (lane 7). Acid gel Northern analysis was performed. Arrows indicate aminoacylated tRNAs, and arrowheads indicate nonaminoacylated tRNAs.
tRNAMet isolated from W303 cells under acidic conditions (Fig. 2, lane 2) is apparently completely aminoacylated. Even though greater than 30% of the total cellular tRNA is located in the nucleus, all detectable tRNAs in nup116Δ cells are aminoacylated (Fig. 2, lane 3). Therefore, assessment of steady-state levels of tRNA indicates that aminoacylation can occur in the nuclear compartment in yeast.
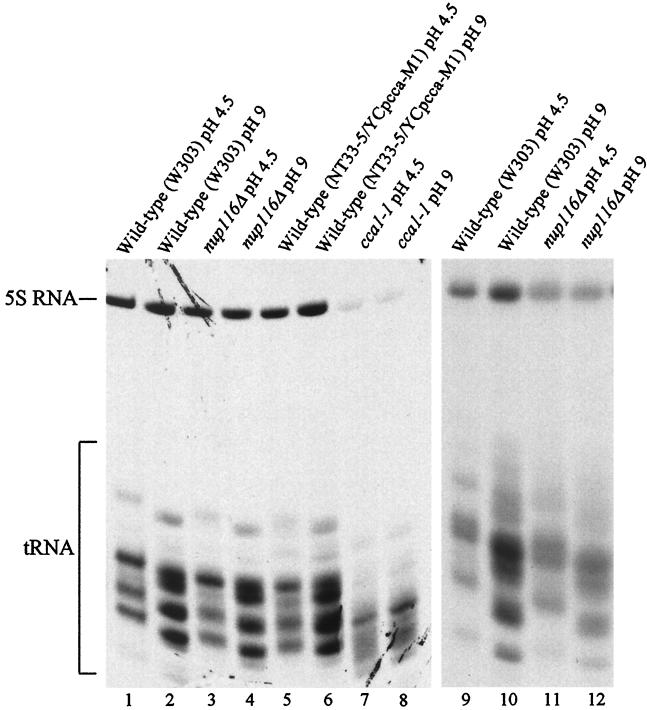
To determine whether other newly synthesized tRNAs that accumulated in the nucleus are aminoacylated, we analyzed tRNAs labeled in vivo under conditions in which the nuclear pores were defective (16, 26). Under these conditions, considerably more than one-third, and perhaps nearly all, of the newly synthesized tRNAs should be located in the nucleus. [3H]uracil was added to the cells after a 10-min (Fig. 3, lanes 1–8) or a 1-h (Fig. 3, lanes 9–12) incubation at 37°C. Labeling proceeded for 1 h before harvesting cells and preparation of RNA by using procedures that would inhibit charging in cellular extracts. To ensure that charging did not occur during extraction, we also added an unlabeled 10-fold excess of nonaminoacylated competitor tRNA to the cells (Fig. 3, lanes 9–12) before processing.
Figure 3.
Aminoacylation status of newly synthesized tRNA. Cells were incubated at 37°C for either 10 min (lanes 1–8) or 1 h (lanes 9–12) before addition of [3H]uracil. Aminoacylated (lanes 1, 3, 5, 7, 9, and 11) and nonaminoacylated (lanes 2, 4, 6, 8, 10, and 12) tRNAs were isolated from W303 wild-type cells (lanes 1, 2, 9, and 10); SWY27, nup116Δ (lanes 3, 4, 11, and 12); NT33-5/YCpcca-M1, wild-type (lanes 5 and 6); and NT33-5/YCp50, cca1-1 (lanes 7 and 8; ref. 19) in the absence (lanes 1–8) or presence (lanes 9–12) of a 10-fold excess of nonaminoacylated yeast tRNA. RNAs were resolved on an acidic polyacrylamide gel and detected by fluorography.
The population of 3H-labeled tRNAs isolated under acidic conditions from wild-type strains migrated more slowly than the corresponding tRNAs isolated under basic conditions, indicating that the majority of newly made cytosolic tRNAs were aminoacylated (Fig. 3, lanes 1, 2, 9, and 10). To ensure that the different mobilities result from differences in aminoacylation, we also isolated RNA from yeast cells with the ts cca1-1 mutation (strain NT33-5/YCp50; ref. 27). At the nonpermissive temperature, these cells accumulate tRNAs that cannot be aminoacylated, because they lack the three 3′ terminal CCA nucleotides (19). As expected, the gel mobilities of tRNAs from cca1-1 cells were faster than those of the wild-type cells because of the lack of the 3′ terminal nucleotides. Also as expected, tRNAs from cca1-1 had the same mobility whether isolated at pH 4.5 or pH 9 (Fig. 3, lanes 7 and 8), confirming that the mobility differences are likely to be the result of aminoacylation; tRNAs isolated from the corresponding wild-type strain (NT33-5/YCpcca-M1) had mobilities similar to those observed for the newly synthesized tRNAs of strain W303 (Fig. 3, compare lanes 5 and 6 with 1 and 2).
The mobility pattern for tRNAs isolated from nup116Δ cells was indistinguishable from that of the parent (Fig. 3, compare lanes 1 and 2 with 3 and 4), even when excess competitor tRNAs were added before cell breakage (Fig. 3, compare lanes 9 and 10 with 11 and 12). The data strongly suggest that most newly synthesized tRNAs, the majority of which should be confined within the nucleus, are aminoacylated.
Our data (Figs. 2 and 3) provide strong evidence for tRNA aminoacylation within the nucleus of S. cerevisiae. Thus, nuclear aminoacylation of tRNAs is not limited to Xenopus oocytes, which are storage cells blocked in meiosis, but also occurs in mitotically growing yeast cells containing endogenous levels of tRNAs. How aminoacyl-tRNA synthetases gain access to the nuclear interior and how their nucleus/cytosol distribution is regulated are challenging questions for future studies (28).
Effects of Aminoacyl-tRNA Synthetase Mutants On Nuclear Export of Cognate and Noncognate tRNAs.
If aminoacylation is important for tRNA nuclear export, then mutations affecting tRNA charging should inhibit export. Aminoacylation requires CCA at the 3′ end of tRNAs, and cells with a ts cca1-1 allele accumulate uncharged tRNAs (Fig. 3, lanes 7 and 8). We employed FISH to locate tRNAs in the cca1-1 mutant and found that, consistent with earlier studies (13), tRNA export, but not poly(A) RNA export, was inhibited (data not shown). However, because tRNAs lacking 3′ CCA are also inefficiently recognized by exportin-t (9, 14, 15), it is not possible to distinguish whether inhibition of tRNA export results from lack of aminoacylation or lack of recognition by the yeast exportin-t homologue, Los1p.
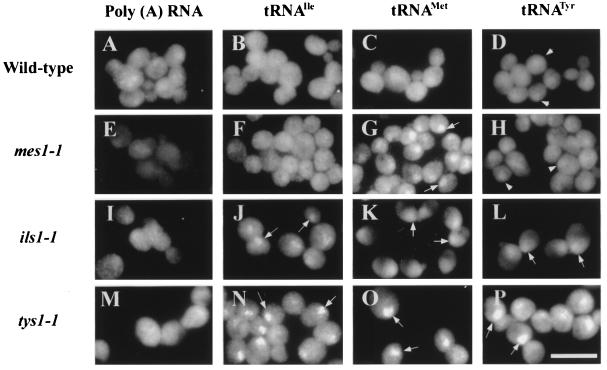
To test the role of tRNA aminoacylation in tRNA directly, we monitored tRNA export in strains with ts mutations of three different aminoacyl-tRNA synthetases. These are mes1-1 (strain ts19:3:4), which encodes a ts methionyl-tRNA synthetase (18); ils1-1 (strain ts341), which encodes a ts isoleucine-tRNA synthetase (17); and tys1-1 (strain ts2), which encodes a ts tyrosyl-tRNA synthetase. The latter mutant was identified by using FISH to screen a collection of ts mutants (29) in an attempt to identify genes specifically involved in nuclear export of tRNA. The identified mutant (ts2) affected tRNA nuclear export but not poly(A) nuclear export (Fig. 4). Genetic analyses showed that nuclear accumulation of tRNA cosegregated with the ts2 lesion. Subcloning of a genomic DNA library inserted in a low-copy centromere-containing plasmid that complements the ts defect revealed that the ORF responsible was TYS1, the gene encoding tyrosine tRNA synthetase (Fig. 5).
Figure 4.
Defect in tRNA nuclear export in aminoacyl-tRNA synthetase mutants. A364A (A–D), mes1-1 (E–H), ils1-1 (I–L), and tys1-1 (M–P) were grown at 23°C, and log phase cells were shifted to 37°C for 2 h. FISH was used to detect poly(A) RNA (A, E, I, and M), tRNAIle (B, F, J, and N), tRNAMet (C, G, K, and O), and tRNATyr (D, H, L, and P). Arrows show locations of nuclei. Arrowheads indicate signal from nucleus-located intron-containing pre-tRNATyr. (Bar = 10 μm.)
Figure 5.
(Upper) Restriction map of TYS1 and flanking region. Numbers 1, 2, and 3 are reading frames from one direction, and −1, −2, and −3 are reading frames from the opposite direction. The half bars represent ATG codons, and the full bars represent stop codons. A segment of UBR1, which codes for ubiquitin-protein ligase, is present in the DNA fragment. TYS1 encodes tyrosyl-tRNA synthetase. TFG1 encodes the large subunit of transcription factor TFIIF. HGH1 encodes HMG1/2 homologue. BUB1 encodes a serine/threonine protein kinase. Abbreviations for restriction enzymes are as follows: B, BamHI; S, SacI; Sa, SalI. (Lower) Mapping of the TYS1 gene. Complete rescue of the growth of tys1-1 at 37°C is shown by + and nonrescue by − for each fragment.
Each synthetase mutant affects nuclear export of its cognate tRNA. tRNAMet accumulates in nuclei of mes1-1 cells (Fig. 4G); tRNAIle accumulates in nuclei of ils1-1 cells (Fig. 4J); and tRNATyr accumulates in nuclei of tys1-1 cells (Fig. 4P). As expected, the mutants and parent cells are not defective in nuclear export of poly(A)-containing mRNA (Fig. 4 A, E, I, and M). Therefore, aminoacylation is important for tRNA nuclear export.
Mutation of a given aminoacyl-tRNA synthetase would not be expected to block nuclear export of noncognate tRNAs. However, because we identified the tys1-1 mutant employing a probe for tRNAIleAAU, we suspected that this expectation may not always hold true. To test this suspicion, we studied the distributions of tRNATyr, tRNAIleAAU, and tRNAMet with noncognate synthetase mutants. tRNAIleAAU and tRNATyr were primarily in the cytosol in mes1-1 cells (Fig. 4 F and H), as in wild-type cells (Fig. 4 B and D). mes1-1 cells have a faint nuclear signal for tRNATyr (Fig. 4H). This signal is likely to be due to nucleus-located intron-containing pre-tRNAs, because we showed that probes for mature tRNA species hybridize to intron-containing pre-tRNA (7); consistent with this explanation, wild-type cells also have a weak nuclear tRNATyr signal (Fig. 4D). Nuclear signals are not observed in wild-type cells for tRNAIleAAU and tRNAMet, because they are encoded by genes that lack introns.
In contrast to mes1-1, tRNA nuclear export of all three tRNAs is defective in ils1-1 and tys1-1 cells (Fig. 4 J–L and N–P). Therefore, although the mes1-1 mutation seems to cause a defect in export of only cognate tRNA, mutations of the two other aminoacyl-tRNA synthetases cause nuclear accumulation of noncognate as well as cognate tRNAs.
The reason why inactivation of some aminoacyl-tRNA synthetases affects export of noncognate tRNAs is unclear. Noncognate tRNAs might accumulate in nuclei of ils1-1 and tys1-1 cells if the mutations affected activity of more than one aminoacyl-tRNA synthetase. Although multienzyme complexes of aminoacyl-tRNA synthetases have been reported not to exist in yeast (30, 31), yeast Arc1p interacts physically and genetically with two aminoacyl-tRNA synthetases, and it has homology to a component of the human aminoacyl-tRNA synthetase multienzyme complex (32, 33). However, aminoacylation of tRNAMet in ils1-1 cells was indistinguishable from that in wild-type cells, indicating that the ils1-1 mutation does not affect methionyl-tRNA synthetase activity (Fig. 2, lanes 4 and 5). Similar experiments with tys1-1 cells showed that this mutation also did not affect aminoacylation of noncognate tRNAMet and tRNAIle (data not shown). Thus, nuclear accumulation of noncognate tRNA is unlikely to result from aberrant aminoacylation in the nucleus.
Defects in protein synthesis and subsequent nuclear signaling might account for nuclear accumulation of noncognate tRNAs in the ils1-1 and tys1-1 mutants. Depending on the aminoacylated tRNA pools, depletion of specific tRNAs might have different effects on protein synthesis and tRNA nuclear export. To investigate this possibility, we determined the location of tRNAs in cells with a ts mutation in the PRT1 gene, which causes inhibition of initiation of protein synthesis at the nonpermissive temperature (34). No defect in the nuclear export of tRNAIleAAU, tRNAMet, or tRNATyr was evident in prt1-1 cells (data not shown). These data are in agreement with studies (13) showing that the protein synthesis inhibitor cycloheximide does not block tRNA nuclear export in Xenopus oocytes.
tRNAIleAAU and tRNATyr are abundant tRNAs, but tRNAMet is not (35), suggesting that defects in aminoacylation of an abundant tRNA might raise its nuclear pool enough to saturate the tRNA export machinery, thereby affecting export of noncognate tRNAs. Indeed, it has been shown in Xenopus that elevated nuclear pre-tRNA levels can saturate the splicing machinery (10, 13) and that elevated levels of injected tRNAs can inhibit tRNA export (3). Future studies altering tRNA cellular levels and/or determining the effect of aminoacylation of other abundant vs. rare tRNAs could provide tests for this hypothesis.
Multiple tRNA Export Pathways?
Our studies support the hypothesis that nuclear aminoacylation is important for tRNA nuclear export (13). Multiple paths for tRNA export may exist (for review, see ref. 36). Previously, it has been suggested that diffusion may be one pathway (15). We favor the notion that Los1p and tRNA aminoacylation are components of two major parallel pathways and that each is capable of proofreading. Thus, cells lacking Los1p, the yeast exportin-t, accumulate tRNAs in nuclei (7) and are still viable through use of the alternative pathway. This model is consistent with the synthetic lethal interactions of arc1 and gcd11 (encoding eIF2γ) mutations with los1 mutations (11, 32). Arc1p interacts with two aminoacyl-tRNA synthetases, and eIF2γ interacts with aminoacylated tRNAs. If Los1p and aminoacyl-tRNA synthetases are components of parallel pathways, then mutations in either that do not cause lethality by themselves could cause lethality when in combination. The model can also account for the data obtained from higher eukaryotes, where it is observed that neither prevention of aminoacylation (13–15) nor sequestration of exportin-t (14, 15) totally blocks tRNA nuclear export.
Acknowledgments
We thank Elsebet Lund for continuous valuable scientific interactions, R. Wek for stimulating ideas, and J. Abelson for the ts collection of yeast. J. Dahlberg, W.-Q. Feng, J. Hengst, J. Hopper, R. Keil, L. Palmer, G. Peng, E. Phizicky, A. Sil, and D. Stanford provided comments on the manuscript. This work was supported by a Public Health Service grant from the National Institutes of Health to A.K.H.
Abbreviations
- ts
temperature-sensitive
- FISH
fluorescence in situ hybridization
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Zasloff M. Proc Natl Acad Sci USA. 1983;80:6436–6440. doi: 10.1073/pnas.80.21.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohno M, Fornerod M, Mattaj I W. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 3.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weis K. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 5.Hopper A K, Martin N C. In: The Molecular and Cellular Biology of Yeast Saccharomyces. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 99–142. [Google Scholar]
- 6.Izaurralde E, Adam S. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar S, Hopper A K. Mol Biol Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kutay U, Lipowsky G, Izaurralde E, Bischoff F R, Schwarzmaier P, Hartmann E, Gorlich D. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- 10.Arts G J, Fornerod M, Mattaj I W. Curr Biol. 1998;8:305–314. doi: 10.1016/s0960-9822(98)70130-7. [DOI] [PubMed] [Google Scholar]
- 11.Hellmuth K, Lau D M, Bischoff F R, Kunzler M, Hurt E, Simos G. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurt D J, Wang S S, Lin Y H, Hopper A K. Mol Cell Biol. 1987;7:1208–1216. doi: 10.1128/mcb.7.3.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lund E, Dahlberg J E. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- 14.Arts G J, Kuersten S, Romby P, Ehresmann B, Mattaj I W. EMBO J. 1998;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipowsky G, Bischoff F R, Izaurralde E, Kutay U, Schafer S, Gross H J, Beier H, Gorlich D. RNA. 1999;5:539–549. doi: 10.1017/s1355838299982134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wente S R, Blobel G. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartwell L H, McLaughlin C S. Proc Natl Acad Sci USA. 1968;59:422–428. doi: 10.1073/pnas.59.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLaughlin C S, Hartwell L H. Genetics. 1969;61:557–566. doi: 10.1093/genetics/61.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfe C L, Hopper A K, Martin N C. J Biol Chem. 1996;271:4679–4686. doi: 10.1074/jbc.271.9.4679. [DOI] [PubMed] [Google Scholar]
- 20.Kuo C L, Campbell J L. Mol Cell Biol. 1983;3:1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopper A K, Schultz L D, Shapiro R A. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- 22.Varshney U, Lee C P, RajBhandary U L. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 23.Keith G, Gangloff J, Dirheimer G. Biochimie. 1971;53:661–669. doi: 10.1016/s0300-9084(71)80022-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang S S, Hopper A K. Mol Cell Biol. 1988;8:5140–5149. doi: 10.1128/mcb.8.12.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hentzen D, Mandel P, Garel J P. Biochim Biophys Acta. 1972;281:228–232. doi: 10.1016/0005-2787(72)90174-8. [DOI] [PubMed] [Google Scholar]
- 26.Hopper A K, Banks F, Evangelidis V. Cell. 1978;14:211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 27.Aebi M, Kirchner G, Chen J Y, Vijayraghavan U, Jacobson A, Martin N C, Abelson J. J Biol Chem. 1990;265:16216–16220. [PubMed] [Google Scholar]
- 28.Schimmel P, Wang C C. Trends Biochem Sci. 1999;24:127–128. doi: 10.1016/s0968-0004(99)01369-9. [DOI] [PubMed] [Google Scholar]
- 29.Vijayraghavan U, Company M, Abelson J. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 30.Kerjan P, Cerini C, Semeriva M, Mirande M. Biochim Biophys Acta. 1994;1199:293–297. doi: 10.1016/0304-4165(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 31.Mirande M. Prog Nucleic Acid Res Mol Biol. 1991;40:95–142. doi: 10.1016/s0079-6603(08)60840-5. [DOI] [PubMed] [Google Scholar]
- 32.Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt E C. EMBO J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- 33.Simos G, Segref A, Fasiolo F, Hellmuth K, Shevchenko A, Mann M, Hurt E C. EMBO J. 1996;15:5437–5448. [PMC free article] [PubMed] [Google Scholar]
- 34.Hartwell L H, McLaughlin C S. Proc Natl Acad Sci USA. 1969;62:468–474. doi: 10.1073/pnas.62.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Percudani R, Pavesi A, Ottonello S. J Mol Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- 36.Simos G, Hurt E. Curr Biol. 1999;9:R238–R241. doi: 10.1016/s0960-9822(99)80152-3. [DOI] [PubMed] [Google Scholar]