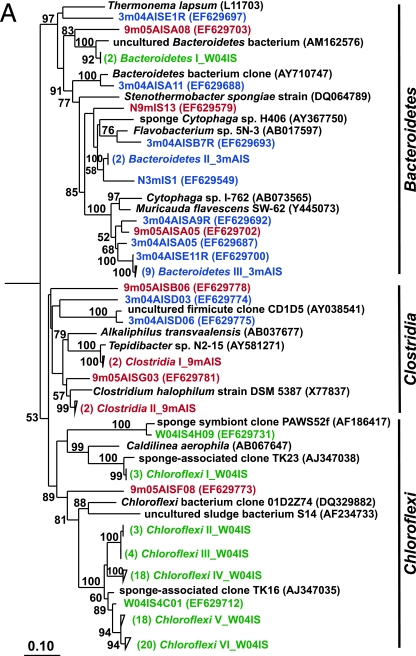
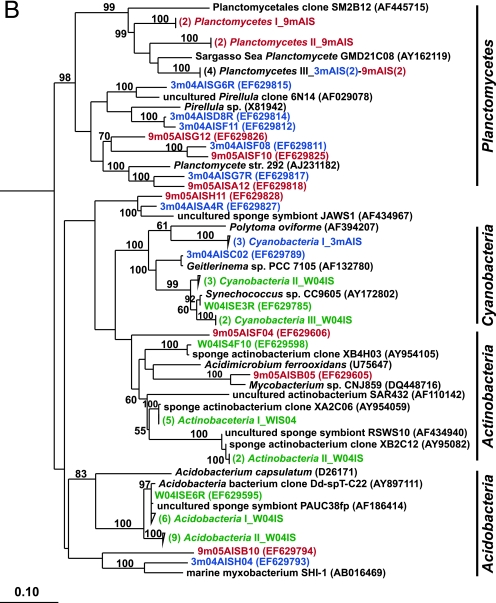
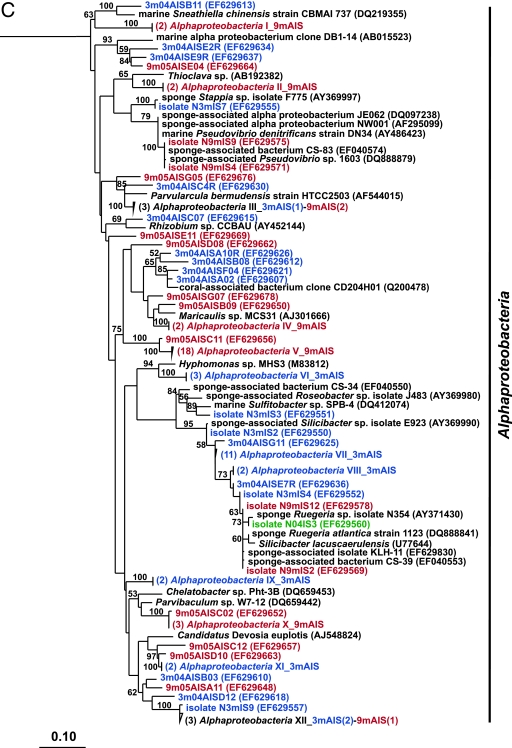
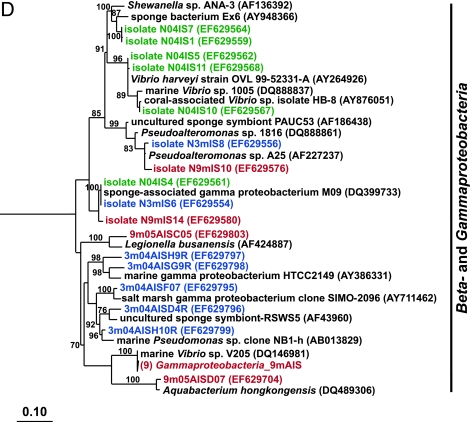
FIG. 2.
Rooted neighbor-joining phylogenetic trees of partial 16S rRNA genes of isolates and clones in the Bacteroidetes, Chloroflexi, and Clostridia (A); Acidobacteria, Actinobacteria, Cyanobacteria, and Planctomycetes (B); Alphaproteobacteria (C); and Beta- and Gammaproteobacteria (D). Isolates and clones were recovered from I. strobilina sponges collected from the wild (prefixed N04IS for isolates and W04IS for clones and shown in green) and maintained for 3 months (prefixed N3mIS for isolates and 3m04AIS for clones and shown in blue) and 9 months (prefixed N9mIS for isolates and 9m05AIS for clones and shown in red) in the aquaculture system. Bootstrap confidence values of >50% are shown at the nodes. The polygons represent clones that are ≥98% similar. The composition of each of these groups is shown in Table S1 in the supplemental material. The numbers listed in bold before the group names indicate the numbers of clones. Thermotoga maritima was used as an outgroup. The scale bar indicates 0.10 substitution per nucleotide position. Reference sequences are shown in bold, with GenBank accession numbers listed after each sequence name.