Abstract
The extent of knowledge regarding the diversity of globally distributed Ehrlichia canis strains has been limited to information gained from a few evolutionarily conserved genes. In this study, E. canis strains from the United States (strain Jake [US]), Brazil (strain São Paulo [BR]), and Israel (strain 611 [IS] and Ranana [IS-R]) were used to examine the antigenic and genetic diversities of four well-characterized major immunoreactive protein genes/proteins. gp36 and gp200 were the most divergent genes, and nucleotide substitutions in the gp36 tandem repeat region of the IS strain, but not the IS-R strain, resulted in two amino acid differences (S→P and P→T) in each nine-amino-acid repeat (epitope-containing region). DNA sequences of gp19 and gp140 were completely conserved in the US and BR strains, but differences were found in the Israeli strains, including two fewer tandem repeats in gp140 and a single amino acid substitution in gp19 from the IS strain. E. canis whole-cell lysates from each isolate were examined by Western immunoblotting using sera from naturally infected dogs from each country, and four major immunoreactive proteins (gp19, gp36, gp140, and gp200) were identified in each strain using protein-specific antisera. The US and BR strains exhibited highly conserved immunoreactive protein profiles, while some differences were identified in the IS strain. Sera from naturally infected Israeli dogs confirmed gene sequencing information, which demonstrated two distinct E. canis strains, defined by the gp36 gene. Conversely, gp19 was strongly reactive and present in all E. canis isolates. gp140 and gp200 were also present in all strains, although gp140 in the IS strain had two fewer tandem repeats and exhibited a smaller mass.
Ehrlichia canis is a globally distributed, tick-transmitted, obligately intracellular bacterium that is the primary etiological agent of canine monocytic ehrlichiosis and has been identified as being the cause of human ehrlichiosis in patients from Venezuela (38, 39). Rickettsiosis in dogs caused by E. canis was first reported in 1935 in Algeria and was later reported in southern India and other parts of Africa in the 1940s (9, 31). Subsequently, E. canis was relatively unrecognized until it was associated with outbreaks of canine tropical pancytopenia in Singapore and Malaysia from 1963 to 1968 (51) and was identified as being the cause of an epizootic of canine tropical pancytopenia in U.S. military dogs stationed in Vietnam in late 1968 (17, 36). E. canis infections have since been well documented in the United States, Israel, Brazil, and Vietnam (1, 3, 12, 16, 20-22, 36, 49), and serologic and/or molecular evidence of infection in temperate regions where Rhipicephalus sanguineus is commonly found, including Central and South America, the Caribbean, parts of Africa, southern Europe, and southeast Asia, has also been reported (2, 5-8, 15, 18, 19, 23, 32, 33, 41, 42, 44, 50).
The development of globally useful serologically and molecularly based diagnostics as well as effective vaccines for canine monocytic ehrlichiosis is dependent on an understanding of the genetic diversity of E. canis, particularly with respect to major immunoreactive proteins. Molecular characterization of evolutionarily conserved genes such as 16S rRNA has provided little information on strain diversity and suggests a high level of conservation (39, 40, 43, 47, 48). Similarly, the immunoreactive major outer membrane proteins p28 and p30 in U.S. and Venezuelan strains of E. canis appear to be highly conserved (13, 29, 30, 46), an observation that was extended to characterized E. canis strains from six human patients from Venezuela (38). Other genes such as the thio-oxidoreductase gene (dsb) and gltA were also found to be conserved in geographically dispersed strains (23, 32).
The genome of E. canis has been sequenced, and a small group of acidic tandem repeat- and ankyrin repeat-containing proteins associated with host-pathogen interactions were identified (24). Several of these proteins are considered major immunoreactive proteins and have been well studied, including gp200, gp140, gp36, and gp19 (11, 25, 26, 28, 53). E. canis gp36 is an acidic serine-rich protein that contains a major antibody epitope in the tandem repeat region (11). Examination of the gp36 gene in U.S., Brazilian, and Cameroonian strains of E. canis identified variations in the numbers of tandem repeats and nucleic acid changes that resulted in four amino acid substitutions (10). However, the diversities of other major immunoreactive E. canis proteins in globally dispersed strains are not known. A homogeneous pattern of proteins reacting with E. canis dog sera from the United States, France, Israel, and the Virgin Islands by immunoblotting was previously reported (14). However, differences in protein reactivity were noted with sera collected from dogs from Italy and Zimbabwe, suggesting the potential for diversity in the antigenic composition of E. canis strains in these countries (14).
The objective of this study was to determine the genetic and antigenic diversities of proteins subject to immune pressure in globally dispersed strains of E. canis. Four major immunoreactive protein genes (gp200, gp140, gp36, and gp19) were sequenced from each strain, and immunoblotting profiles for E. canis whole-cell lysates were compared. Strains from the United States and Brazil exhibited homogeneous immunoblotting patterns compared to that of the strain from Israel. Sequencing of four major immunoreactive protein genes demonstrated that U.S. and Brazilian strains were highly similar and that strains from Israel were the more divergent.
MATERIALS AND METHODS
Ehrlichia canis strains and propagation.
Ehrlichia canis strains used in this study originated from the United States (strain Jake [US]), Israel (strain 611 [IS]), and Brazil (strain São Paulo [BR]). DNA was also obtained from an Israeli dog (Israeli strain Ranana [IS-R]) naturally infected with E. canis for comparison. E. canis strains (US, IS, and BR) were propagated in DH82 cells (canine histiocyte) with minimal essential medium (Gibco, Grand Island, NY) supplemented with 5% fetal bovine serum (HyClone, Logan, UT), 1% HEPES (Sigma Chemical Co., St. Louis, MO), 1% sodium pyruvate (Sigma), and 1% nonessential amino acids (Sigma). The IS strain in J774 cells (murine) was provided to our laboratory; however, cell-free ehrlichiae from these cultures were used to infect DH82 cells for the antigen used in this study. Infected cells were harvested when morulae were observed in all cells. Cells were pelleted (5,000 × g for 15 min), resuspended in phosphate-buffered saline (PBS), and sonicated twice (40 Hz) for 10 s, and large cell debris was pelleted by centrifugation (1,500 × g for 10 min) at 4°C. The supernatant containing cell-free ehrlichiae was centrifuged (10,000 × g for 15 min) at 4°C. The pellet was then washed once in PBS, pelleted (10,000 × g for 15 min) at 4°C, and resuspended in PBS. The suspension containing bacteria was frozen at −80°C and utilized as an antigen and DNA source. The protein concentration of purified E. canis antigen was determined using the BCA protein assay (Pierce Biotechnology, Rockford, IL).
PCR amplification and cloning of major immunoreactive protein genes.
E. canis genomic DNA was extracted from purified antigen using a commercial kit according to the manufacturer's protocol (MasterPure Complete DNA and RNA purification kit; Epicentre, Madison, WI). The primers used for the amplification of E. canis genes (gp19, gp36, gp140, and gp200) (Table 1) were designed using primer design software (PrimerSelect; DNASTAR, Madison, WI) and E. canis genome sequence information (Integrated Microbial Genomes system; United States Department of Energy, Joint Genome Institute, Walnut Creek, CA).
TABLE 1.
Primers used for the amplification of the gp19, gp36, gp140, and gp200 genes from the US, BR, and IS E. canis strains
| Gene | Forward primer | Sequence of forward primer (5′-3′) | Reverse primer | Sequence of reverse primer (5′-3′) | Amplicon size (bp) |
|---|---|---|---|---|---|
| p19 | p19F | AAAATTAGTGTTGTGGTTATG | p19R | TTTTACGCTTGCTGAAT | 492 |
| p36 | p36F-L | GGAATGATTTATTAAAAAAGTTTGAC | p36R-L | GATCGTTGGATGTTGG | 2,080 |
| p140 | p140F | ATGGATATTGATAACAATAATGTGACTAC | p140R | TATTAAATCAACTGTTTCTTTGTTAGT | 2,061 |
| p200 | p200F | TTTGCCATTCAGGAACATCG | p200R | TGCACCTCATATCCAACTAGAAACAC | 4,597 |
The E. canis gp19 gene (Ecaj_0113) was amplified with primers (Table 1) that target intergenic regions (∼50 bp upstream and ∼9 bp downstream) flanking the gene. The gp19 gene was amplified by PCR using Hot master mix (Eppendorf, Westbury, NY) with the following thermal cycling protocol: 95°C for 4 min and 30 cycles at 95°C for 30 s, 47°C for 30 s, and 72°C for 1 min, followed by a 72°C extension step for 7 min. The E. canis gp36 gene (Ecaj_0109) was amplified using primers (Table 1) that targeted highly conserved genes (upstream, Ecaj_ 0108 [MerR transcriptional regulator]; downstream, Ecaj_0110 [tryptophanyl tRNA synthase]) flanking the gp36 gene. PCR was performed as described above for the gp19 gene except that Platinum Taq DNA Polymerase High Fidelity (Invitrogen; Carlsbad, CA) was used with an annealing temperature of 55°C and an extension step at 72°C for 3 min. E. canis gp140 (Ecaj_0017) was amplified using primers (Table 1) located within the open reading frame (forward, bases 1 to 29; reverse, bases 2034 to 2061). PCR was performed under the conditions described above for the gp36 gene except that an annealing temperature of 57°C for 30 s and an extension step at 72°C for 1.5 min were used. The gp200 gene (Ecaj_0365) was amplified using primers (Table 1) targeting the intergenic region (∼250 bp upstream and ∼20 bp downstream) flanking the gene. The gene was amplified using conditions described above for the gp36 gene except that an annealing temperature of 61°C and an extension step at 72°C for 5 min were used.
PCR amplicons for all four genes were separated and visualized by agarose gel electrophoresis (1.2% FlashGel DNA system; Lonza, Walkersville, MD). The gp19 amplicon was purified using a purification kit (ExoSAP-IT; USB Corp., Cleveland, OH) sequenced directly using the same primers. All other PCR amplicons were cloned directly into universal TOPO TA sequencing vectors (Invitrogen), and plasmids were purified using a plasmid purification kit (High Pure plasmid isolation kit; Roche, Indianapolis, IN) and sequenced using primers supplied with the vector.
DNA sequencing.
PCR amplicons and plasmids were sequenced with an ABI Prism 377XL DNA sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA) at the University of Texas Medical Branch Protein Chemistry Core Laboratory.
Cloning and expression of recombinant gp36 from the IS and US strains.
The gp36 gene from the IS strain (810 bp) was amplified by PCR using forward primer 5′-ATG CTA TTT ATA CTA ATG GGT TAT TG-3′ and reverse primer 5′-CAG GGT AAG CTG AGT ATA TAA ATC-3′ with IS strain DNA as the template with the following thermal cycling protocol: 94°C for 30s, 55°C for 30s, and an extension step at 72°C for 1 min. The PCR product was cloned into the pBAD/Thio fusion vector (Invitrogen), and recombinant proteins (from the IS and US strains) were expressed and purified as previously described (11).
Gel electrophoresis and Western immunoblotting.
Purified E. canis whole-cell lysates (5 μg/well) were solubilized in LDS sample buffer (NuPAGE; Invitrogen) and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using Novex 4 to 12% Bis-Tris gels (15 wells) (NuPAGE; Invitrogen) and morpholinepropanesulfonic acid (MOPS) running buffer (NuPAGE; Invitrogen). Separated lysates were transferred onto pure nitrocellulose (Protran BA85, with a 0.45-μm pore size; Whatman, Florham Park, NJ) using a semidry transfer apparatus (Bio-Rad, Hercules, CA). Anti-E. canis sera (1:2,000 for U.S. dog 02160, 1:1,000 for U.S. dog 04283, 1:1,000 for U.S. dog 20699, 1:1,000 for dog ZOC, 1:1,000 for Israeli dogs 53, 18, 17, and 1, 1:500 for Brazilian dog 157, 1:1,000 for Brazilian dog 37, 1:800 for Brazilian dog 42, and 1:800 for Brazilian dog 45) used for Western immunoblots were obtained from dogs naturally infected with E. canis. Rabbit anti-recombinant protein sera (1:5,000 of gp19, 1:500 for gp36, 1:200 for gp140, and 1:100 for gp200) were used to identify native E. canis immunoreactive proteins in whole-cell lysates from each strain. Western immunoblotting was performed as previously described (25).
Major immunoreactive protein-specific antisera.
Antisera specific for gp200, gp140, gp36, and gp19 were produced in rabbits as previously described (11, 27, 28, 53).
Sequence analysis.
Nucleic acid and amino acid alignments (using the Clustal W algorithm), percent identities, and phylogenetic relationships were determined with MegAlign (Lasergene v5.08; DNAStar, Madison, WI).
Nucleotide sequence accession numbers.
Gene sequences for genes sequenced in this study for E. canis (strain Jake [US]) were previously available in the GenBank database (accession numbers DQ085427 for gp36, DQ858221 for gp19, AF252298 for gp200, and AF112369 for gp140). The E. canis (strain São Paulo [BR]) gp36 gene was also available in the GenBank database (accession number DQ146154). The following accession numbers were assigned to genes from the E. canis BR strain sequenced in this study: EU118960 for gp19, EU118964 for gp140, and EF636664 for gp200. The E. canis genes from the IS strain (strain 611) were assigned the following accession numbers: EU118959 for gp19, EF636663 for gp36, EU118963 for gp140, and EF636665 for gp200. Gene sequences amplified from the strain from the Israeli dog (IS-R) naturally infected with E. canis were assigned the following accession numbers: EU118958 for gp19, EU118961 for gp36, and EU118962 for gp140.
RESULTS
Diverse immunoreactive proteins (gp36 and gp200).
Major immunoreactive protein gp36 and gp200 genes were amplified and sequenced from the three isolates (US, BR, and IS) and one blood sample (gp36 only, as gp200 could not be amplified from blood) from an Israeli dog (IS-R) naturally infected with E. canis (Fig. 1 and 2). gp36 was the most divergent, with amino acid identities ranging from 81.5% to 91.7%. E. canis gp36 (US strain) had 12 tandem repeats, but six additional repeats were found in the BR strain, which decreased the overall percent identity (81.5%). However, only eight nucleotide differences (eight amino acid changes) were found between the gp36 genes from the US and BR strains: four of those were located in the C terminus (the last 15 amino acids). Conversely, substantial divergence in the gp36 gene from the IS strain was found. Two nucleotides of the sequence that encodes the nine-amino-acid repeat differed, resulting in two amino acid substitutions that were noted only in the IS strain. The IS strain had 11 tandem repeats, and the IS-R strain had 10. The repeat region of the US, BR, and IS-R strains were identical in sequence but differed in the numbers of repeats (12, 18, and 10, respectively). The domain with the highest level of divergence in gp36 was found in the C-terminal region (the last 18 amino acids). The IS strain exhibited the least identity (33%) with the US strain in this region, while the BR and IS-R strains had a higher level of identity with the US strain.
FIG. 1.
Clustal alignment of the gp36 amino acid sequence of E. canis strains (US, BR, IS, and IS-R strains) from three continents. Boxed amino acids represent residues divergent from the US strain sequence, and a dash represents a gap. Single tandem repeat units are identified with a bar.
FIG. 2.
Clustal alignment of the gp200 amino acid sequence of E. canis strains (US, BR, and IS strains) from three continents. Boxed amino acids represent residues divergent from the US strain sequence, and a dash represents a gap.
The gp200 proteins from the US and BR strains exhibited a high amino acid identity (99.6%), and the IS strain had lower identity (94.3%). There were four amino acid differences between the US and BR gp200 proteins, including one insertion (position 1360). The IS strain exhibited numerous amino acid changes, with a higher frequency of changes (47%) found in a 325-amino-acid stretch located in the C-terminal region (amino acids 950 to 1275) of the protein.
Conserved immunoreactive protein (gp19 and gp140) genes.
Major immunoreactive protein gp19 and gp140 genes were amplified and sequenced from three isolates (the US, BR, and IS strains) and one blood sample from an Israeli dog (IS-R) naturally infected with E. canis. The US and BR strains had identical gp19 and gp140 gene sequences. The gp19 gene of the IS strain had one nucleotide substitution (position 104) that resulted in a single amino acid change (Glu to Gly at position 35) in the epitope-containing region that was previously reported (28). Interestingly, the gp19 gene amplified from the Israeli dog naturally infected with E. canis (IS-R) was identical to those from the US and BR strains. High degrees of overall nucleic acid and amino acid conservations were observed in gp19 (99.99% to 100% identity) in geographically dispersed strains.
The gp140 gene of the IS strain had two fewer tandem repeats (12 [there were 14 in the US strain]) but had nine nucleotide substitutions that resulted in nine amino acid changes, seven that were localized to 2 of the 12 tandem repeats (Fig. 3). The IS-R strain had the same number of tandem repeats as the IS strain and had identical amino acid changes in seven locations. However, eight additional amino acid changes (in the repeat region) unique to the IS-R strain compared to the IS strain were noted (Fig. 4). High degrees of nucleic acid and amino acid conservations in gp140 (99.8% to 100% identity) were observed in geographically dispersed strains.
FIG. 3.
Clustal alignment of the gp140 amino acid sequence of E. canis strains (US, BR, IS, and IS-R strains) from three continents. Boxed amino acids represent residues divergent from the US strain sequence, and a dash represents a gap. Single tandem repeat units are identified with a bar.
FIG. 4.
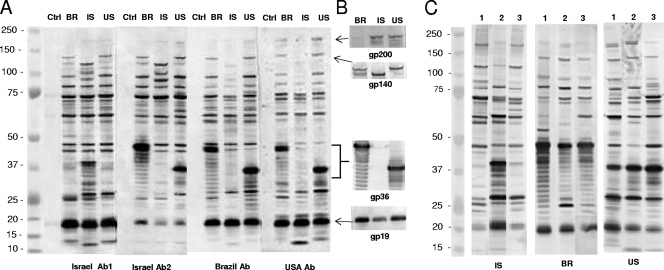
(A) Western immunoblot of E. canis whole-cell lysates (BR, US, and IS strains) and uninfected DH82 cell lysates (control [Ctrl]) probed with sera from dogs naturally infected with E. canis from each respective country (Israeli dogs 18 and 53, Brazilian dog 157, and U.S. dog 02160). (B) E. canis whole-cell lysates (BR, IS, and US strains) probed with rabbit anti-US gp200-, gp140-, gp36-, and gp19-specific sera, respectively. (C) E. canis whole-cell lysates from the IS, BR, and US strains reacted with three sera from dogs naturally infected with E. canis from the United States (dogs 04283 [lane 1], 20699 [lane 2], and ZOC [lane 3]), Brazil (dogs 37 [lane 1], 42 [lane 2], and 45 [lane 3]), and Israel (dogs 17 [lane 1], 18 [lane 2], and 1 [lane 3]). Ab, antibody.
Western immunoblotting.
E. canis whole-cell lysates (from the US, BR, and IS strains) were reacted with sera obtained from dogs naturally infected with E. canis in each respective country (Fig. 4A). The reactivities of all sera with each lysate (from the US, BR, and IS strains) were relatively homogeneous and most consistent in the masses of >45 kDa and <100 kDa. The most notable differences were observed in known major immunoreactive proteins, including gp36 and gp140. gp36 from the BR strain was substantially larger than gp36 from the US strain due to six additional repeat units (Fig. 1 and 4A). Two types of immunoreactivity were consistently observed with Israeli dog sera. One type cross-reacted with gp36 from the US and BR strains (Fig. 4A), and the other type reacted with a protein present in the IS strain that was similar in size to gp36 from the US strain, and these sera did not cross-react with gp36 from the US and BR strains (Fig. 4A). gp140 of the IS strain was smaller (two repeats less) than the US and BR strains (Fig. 3) and was strongly recognized in all strains with the Israeli sera, but gp140 from the IS strain was weakly recognized by sera from strains from Brazil and the United States (Fig. 4A). gp19 exhibited the same mass in all strains and was strongly recognized by dog sera from each country (Fig. 4A). gp200 exhibited a similar mass in all strains, and the immunoreactivity of gp200 was more prominent in the whole-cell lysates from the BR and US strains probed with U.S. sera (Fig. 4A). Another notable difference was an unknown protein (∼90 to 95 kDa) in all strains that strongly reacted with sera from Israel among all strains, but the IS protein was weakly recognized by Brazilian and U.S. sera (Fig. 4A).
Anti-US strain gp36 antibody reacted strongly with gp36 from the BR and US strains, but no reactivity was observed with the IS strain. Anti-US strain gp200 and gp19 antibodies reacted with proteins of similar sizes in all strains (Fig. 4B), and anti-US strain gp140 reacted with all strains, although gp140 in the IS strain was smaller in mass (Fig. 4B).
Three sera from dogs naturally infected with E. canis from Israel, Brazil, and the United States were reacted with the respective E. canis whole-cell lysates (from the IS, BR, and US strains) to evaluate the consistency of antigen recognition by different dogs from the same location. Similar antigen reactivity patterns of the sera with each respective E. canis lysate were observed (Fig. 4C). Some minor differences in antigen recognition among dogs from the same region were noted, but immunodominant proteins were consistently recognized by all dogs. One exception worth noting was observed with the sera from Israel. As noted above, a protein similar in size to gp36 was strongly reactive with two of the Israeli dog sera (Israel Ab1), and two Israeli dog sera (Israel Ab2) reacted with gp36 from the US and BR strains, as shown in Fig. 4A and 5.
FIG. 5.
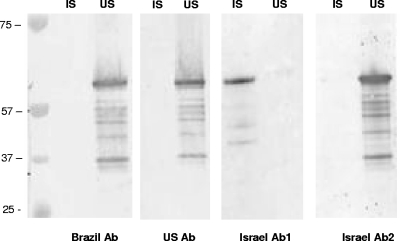
Immunoreactivities of purified recombinant gp36 proteins from the IS (lane 1) and US (lane 2) strains probed with sera from dogs from Brazil, the United States, and Israel naturally infected with E. canis. Ab, antibody.
Immunoreactivity of recombinant gp36 from IS and US strains.
The recombinant gp36 proteins from the IS (strain 611) and US (strain Jake) strains were expressed and reacted with E. canis-infected dog sera from Israel, Brazil, and the United States. The expressed recombinant gp36 from the IS strain exhibited a molecular mass slightly smaller (∼53 kDa, including 16-kDa N- and C-terminal fusion tags) than the recombinant gp36 from the US strain. The recombinant gp36 from the IS strain reacted strongly with sera from an Israeli dog naturally infected with E. canis but not with the recombinant gp36 from the US strain. gp36 from the US strain reacted strongly with sera from the United States and Brazil. Sera from Israeli dogs that recognized gp36 from the IS strain did not cross-react with gp36 from the US strain. In contrast, Israeli sera that recognized gp36 from the US strain did not recognize gp36 from the IS strain (Fig. 5).
DISCUSSION
E. canis is the most widely dispersed Ehrlichia species, yet little information regarding the antigenic variability of the organism is available. Previous studies indicated that some genes including the 16 rRNA, dsb, and p28/p30 genes exhibit a high level of conservation in geographically dispersed strains (13, 32, 38-40, 43, 46). However, one serological comparison also suggested that there is antigenic variability in geographically dispersed E. canis strains (14). We have examined, for the first time, the molecular diversities among four major immunoreactive proteins (gp200, gp140, gp36, and gp19) and compared the reactivities of three (the US, IS, and BR strains) E. canis whole-cell lysates against homologous and heterologous sera from three continents. The major immunoreactive proteins of E. canis elicit a strong immune response, and thus, the genes encoding these proteins may exhibit a higher level of diversity as a result of increased selective immune pressure. Furthermore, two of these genes contain tandem repeats, and variations in the number and sequence of Ehrlichia tandem repeat-containing proteins are well established.
This is the first study in which three globally dispersed strains of E. canis were propagated in a single laboratory in order to closely compare the antigenic profiles under the same conditions and to examine genetic differences in four newly characterized major immunoreactive protein genes in these strains. Previously, sera from various locations were reacted with a single E. canis antigen preparation (US strain) to gain some information regarding antigenic diversity (14). Future studies with more strains from more locations may provide additional insight into the diversity of E. canis; this study does provide important information with regard to E. canis in the three countries included in this investigation. Western immunoblotting of native E. canis lysates reacted with homologous and heterologous sera revealed that the immunoblot pattern of immunoreactive proteins is consistently homogeneous with regard to protein mass and immunoreactivity, suggesting that most of these proteins are conserved among geographically separated strains. These findings are in agreement with a previous study that reported relatively homogeneous immunoblot patterns using sera from different geographic locations (14). However, the former study was limited in that it compared antigen profiles (proteins of <110 kDa) using a single E. canis (strain Florida) whole-cell lysate preparation. Furthermore, consistent and strong recognition of E. canis antigens (>80 kDa) that were reactive by immunoblot in this study was not consistently identified in the previous study using sera originating from the United States (14). This difference is likely due to protein blotting conditions resulting in the more efficient transfer of high-molecular-mass proteins in our study. Furthermore, some notable differences in the molecular masses of immunodominant antigens were also identified in this study. gp19 and gp36 were the most immunodominant antigens in the immunoblots for both the US and BR strains, while proteins in the 28- to 30-kDa range, consistent with the mass of the major outer membrane protein (p28/p30), were present but were less dominant. The immunodominant proteins identified in this study are consistent with those identified in our previous study using the same US strain antigen but with sera from experimentally infected dogs (25).
Antigens that were most visibly different among strains (molecular mass) were some of the proteins specifically examined in this study. The most divergent of the four antigens examined in this study was gp36, a secreted protein that elicits an early antibody response directed at the tandem repeat region and is also differentially expressed on dense-cored ehrlichiae (11). Previous studies reported differences in the numbers of gp36 tandem repeats in E. canis strains (10) as well as the ortholog in Ehrlichia ruminantium (Erum1110) (4), a finding confirmed in this study. Interestingly, there were two types of sera from naturally infected dogs from Israel that were identified based on reactivity to gp36. The first type reacted strongly with gp36 from the US and BR strains, and the second type reacted with a protein in the IS strain with a size similar to that of gp36 from the US strain and consistent with the size of gp36 from the IS strain that was sequenced in this study. This difference can be explained using the gp36 gene sequence information from the IS and IS-R strains. Some dogs appear to be infected with an IS-R strain type, in which the antibody epitope region is identical to that of gp36 from the US strain (11). In contrast, the IS strain, which was propagated in the laboratory and used in the immunoblots, has a divergent gp36, which has two amino acid substitutions (S→P and P→T) in the epitope-containing repeat region. Thus, the serological response to gp36 in the IS antigen preparation is dependent on the strain of E. canis infecting the dogs. Evaluation of four Israeli sera from naturally infected dogs found that half of the sera were specific for gp36 from the IS strain and that half contained antibodies to gp36 from the US strain (IS-R strain). Serological and molecular evidences indicate that there are at least two distinct E. canis strains circulating in Israel. This is in contrast to strains circulating in the United States and Brazil, which appear to be more conserved, as was previously reported (10, 11, 29, 30, 53). Another interesting region of diversity among all strains was in the gp36 carboxy-terminal region immediately downstream of the repeat region. Antibody epitopes have not been identified in this region (11), suggesting that this diversity is not a direct result of humoral immune selection pressure. The divergence of gp36 among the strains examined in this study supports the conclusion that this gene is useful for the molecular genotyping of E. canis strains.
gp200, which is the largest major immunoreactive protein identified in E. canis, had a high level of conservation between the US and BR strains, but substantial diversity was found in the IS strain. gp200 is a secreted nuclear translocated ankyrin repeat-containing protein that has five major species-specific epitopes located primarily in terminal acidic domains (34, 35). The amino acid changes in gp200 from the IS strain were distributed throughout the protein, but a higher frequency of amino acid substitutions was noted in a carboxy-terminal 325-amino-acid domain of the protein. Amino acid substitutions were identified in known gp200 epitopes (35). The carboxy-terminal and internal epitopes had at least two amino acid substitutions; however, only one of the two known amino-terminal epitopes had a single amino acid substitution. gp200 from the IS strain appeared to be less reactive with heterologous sera than with homologous sera, and these substitutions in known epitopes are likely responsible for this difference. Conversely, the conservation of the N-terminal epitopes would result in the recognition of gp200 in all strains, as was demonstrated by immunoblotting.
Two of the immunoreactive proteins (gp19 and gp140) examined in this study were highly conserved. gp140 was previously shown to be identical in strains from the United States (53); however, the conservation of gp140 outside the United States has not been investigated. Consistent with previous findings, gp140 was found to be highly conserved among the US, BR, and IS strains. The IS-R strain was the most divergent, with the most frequent substitution located in the repeat region, where an asparagine was replaced by serine. This substitution was also observed in the IS strain but was more frequent in the IS-R strain. The fact that all of the tandem repeats lacked this substitution suggests that it is a point mutation that is occurring as a result of immune pressure. The repeat region of gp140 does contain a strong antibody epitope in an area containing the substituted amino acid (J. W. McBride, unpublished data). The most notable difference was found in the Israeli strains, which had two fewer tandem repeats. This difference in tandem repeats was also evident by immunoblotting, as gp140 from the IS strain exhibited a smaller molecular mass. Variations in the numbers of tandem repeats have been reported for Ehrlichia chaffeensis gp120, the E. canis gp140 ortholog (45, 52). However, E. chaffeensis gp120 repeat variants have not been associated with differences in pathogenicity (37).
E. canis gp19 is a recently characterized ortholog of E. chaffeensis variable-length PCR target protein and has a single major serine-rich epitope (28). In our previous study, we reported that gp19 was highly conserved among strains from the United States, Mexico, Brazil, and Israel. However, single amino acid changes were noted in the Israeli and Mexican strains, both of which were located in the antibody epitope-containing region (28). In this study, the IS-R strain was found to be identical to the US and BR strains, and the IS strain (611) was confirmed to have a single amino acid substitution at position 35. Although the IS strain has a single amino acid substitution in the epitope-containing region, it was still strongly recognized by homologous and heterologous antisera, suggesting that this change is not critical for epitope recognition. However, selective immune pressure may be responsible for these changes, considering the location. gp19 does elicit an early antibody response and is a dominant antigen on immunoblots. The conservation of this antigen in geographically dispersed E. canis strains suggests that this protein can be useful for immunodiagnostics and vaccines that are widely applicable.
The development of reliable immunodiagnostics and vaccines for canine ehrlichiosis is dependent on an understanding of differences that may exist in geographically dispersed strains of E. canis, particularly with respect to these important genes. Further studies involving additional E. canis strains and more potentially important genes are needed to appreciate the full extent of global diversity of the organism and specific genes that have increased selection pressure. We specifically focused this study on genes with increased selection pressure in order to provide additional insight into the diversity of E. canis strains. This information would also expand our knowledge with regard to the genetic variability in known targets of the host immune response and identify new and useful targets for genotyping of E. canis. Based on the information generated in this study and others, E. canis appears to be more conserved than E. chaffeensis or E. ruminantium, but the variability in the major immunoreactive proteins examined in this study indicates that substantial variability is present among E. canis strains. Furthermore, it is evident that variability in E. canis strains must be a consideration in developing widely applicable diagnostics and vaccines.
Acknowledgments
We thank Osnat Eyal for technical assistance and sequencing of the gp19 gene from the IS-R strain and Xue-jie Yu and David H. Walker for critical review of the manuscript.
This work was supported by the National Institutes of Health (AI 071145-01) and the Clayton Foundation for Research.
Footnotes
Published ahead of print on 14 May 2008.
REFERENCES
- 1.Aguiar, D. M., G. T. Cavalcante, A. Pinter, S. M. Gennari, L. M. Camargo, and M. B. Labruna. 2007. Prevalence of Ehrlichia canis (Rickettsiales: Anaplasmataceae) in dogs and Rhipicephalus sanguineus (Acari: Ixodidae) ticks from Brazil. J. Med. Entomol. 44:126-132. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre, E., A. Sainz, S. Dunner, I. Amusategui, L. Lopez, F. Rodriguez-Franco, I. Luaces, O. Cortes, and M. A. Tesouro. 2004. First isolation and molecular characterization of Ehrlichia canis in Spain. Vet. Parasitol. 125:365-372. [DOI] [PubMed] [Google Scholar]
- 3.Baneth, G., T. Waner, A. Koplah, S. Weinstein, and A. Keysary. 1996. Survey of Ehrlichia canis antibodies among dogs in Israel. Vet. Rec. 138:257-259. [DOI] [PubMed] [Google Scholar]
- 4.Barbet, A. F., W. M. Whitmire, S. M. Kamper, B. H. Simbi, R. R. Ganta, A. L. Moreland, D. M. Mwangi, T. C. McGuire, and S. M. Mahan. 2001. A subset of Cowdria ruminantium genes important for immune recognition and protection. Gene 275:287-298. [DOI] [PubMed] [Google Scholar]
- 5.Brouqui, P., B. Davoust, S. Haddad, E. Vidor, and D. Raoult. 1991. Serological evaluation of Ehrlichia canis infections in military dogs in Africa and Reunion Island. Vet. Microbiol. 26:103-105. [DOI] [PubMed] [Google Scholar]
- 6.Costa, L. M., Jr., K. Rembeck, M. F. Ribeiro, P. Beelitz, K. Pfister, and L. M. Passos. 2007. Sero-prevalence and risk indicators for canine ehrlichiosis in three rural areas of Brazil. Vet. J. 174:673-676. [DOI] [PubMed] [Google Scholar]
- 7.Dagnone, A. S., H. S. de Morais, M. C. Vidotto, F. S. Jojima, and O. Vidotto. 2003. Ehrlichiosis in anemic, thrombocytopenic, or tick-infested dogs from a hospital population in South Brazil. Vet. Parasitol. 117:285-290. [DOI] [PubMed] [Google Scholar]
- 8.Davoust, B., O. Bourry, J. Gomez, L. Lafay, F. Casali, E. Leroy, and D. Parzy. 2006. Surveys on seroprevalence of canine monocytic ehrlichiosis among dogs living in the Ivory Coast and Gabon and evaluation of a quick commercial test kit dot-ELISA. Ann. N. Y. Acad. Sci. 1078:464-469. [DOI] [PubMed] [Google Scholar]
- 9.Donatien, A., and F. Lestoquard. 1935. Existence en Algerie d'une rickettsia du chien. Bull. Soc. Pathol. Exot. 28:418-419. [Google Scholar]
- 10.Doyle, C. K., A. M. Cardenas, D. M. Aguiar, M. B. Labruna, L. M. Ndip, X. J. Yu, and J. W. McBride. 2006. Molecular characterization of E. canis gp36 and E. chaffeensis gp47 tandem repeats among different geographic locations. Ann. N. Y. Acad. Sci. 1063:433-435. [DOI] [PubMed] [Google Scholar]
- 11.Doyle, C. K., K. A. Nethery, V. L. Popov, and J. W. McBride. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing, S. A. 1963. Observations of leukocytic inclusion bodies from dogs infected with Babesia canis. J. Am. Vet. Med. Assoc. 143:503-506. [PubMed] [Google Scholar]
- 13.Felek, S., R. Greene, and Y. Rikihisa. 2003. Transcriptional analysis of p30 major outer membrane protein genes of Ehrlichia canis in naturally infected ticks and sequence analysis of p30-10 of E. canis from diverse geographic regions. J. Clin. Microbiol. 41:886-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hegarty, B. C., M. G. Levy, R. F. Gager, and E. B. Breitschwerdt. 1997. Immunoblot analysis of the immunoglobulin G response to Ehrlichia canis in dogs: an international survey. J. Vet. Diagn. Investig. 9:32-38. [DOI] [PubMed] [Google Scholar]
- 15.Hua, P., M. Yuhai, T. Shide, S. Yang, W. Bohai, and C. Xiangrui. 2000. Canine ehrlichiosis caused simultaneously by Ehrlichia canis and Ehrlichia platys. Microbiol. Immunol. 44:737-739. [DOI] [PubMed] [Google Scholar]
- 16.Huxsoll, D. L., P. K. Hildebrandt, and R. M. Nims. 1970. Tropical canine pancytopenia. J. Am. Vet. Med. Assoc. 157:1627-1632. [PubMed] [Google Scholar]
- 17.Huxsoll, D. L., P. K. Hildebrandt, R. M. Nims, J. A. Ferguson, and J. S. Walker. 1969. Ehrlichia canis—the causative agent of a haemorrhagic disease of dogs? Vet. Rec. 85:587. [DOI] [PubMed] [Google Scholar]
- 18.Inokuma, H., M. Oyamada, B. Davoust, M. Boni, J. Dereure, B. Bucheton, A. Hammad, M. Watanabe, K. Itamoto, M. Okuda, and P. Brouqui. 2006. Epidemiological survey of Ehrlichia canis and related species infection in dogs in eastern Sudan. Ann. N. Y. Acad. Sci. 1078:461-463. [DOI] [PubMed] [Google Scholar]
- 19.Jouret-Gourjault, S., D. Parzy, and B. Davoust. 2006. Experimental infections in dogs with Ehrlichia canis strain Borgo 89. Ann. N. Y. Acad. Sci. 1078:470-475. [DOI] [PubMed] [Google Scholar]
- 20.Kelch, W. J. 1984. The canine ehrlichiosis (tropical canine pancytopenia) epizootic in Vietnam and its implications for the veterinary care of military working dogs. Mil. Med. 149:327-331. [PubMed] [Google Scholar]
- 21.Keysary, A., T. Waner, M. Rosner, C. K. Warner, J. E. Dawson, R. Zass, K. L. Biggie, and S. Harrus. 1996. The first isolation, in vitro propagation, and genetic characterization of Ehrlichia canis in Israel. Vet. Parasitol. 62:331-340. [DOI] [PubMed] [Google Scholar]
- 22.Komnenou, A. A., M. E. Mylonakis, V. Kouti, L. Tendoma, L. Leontides, E. Skountzou, A. Dessiris, A. F. Koutinas, and R. Ofri. 2007. Ocular manifestations of natural canine monocytic ehrlichiosis (Ehrlichia canis): a retrospective study of 90 cases. Vet. Ophthalmol. 10:137-142. [DOI] [PubMed] [Google Scholar]
- 23.Marsilio, F., B. Di Martino, I. Meridiani, and P. Bianciardi. 2006. Direct identification of Ehrlichia canis by a novel polymerase chain reaction method and molecular analysis of the citrate synthase (gltA) gene from various Italian strains. J. Vet. Diagn. Investig. 18:215-217. [DOI] [PubMed] [Google Scholar]
- 24.Mavromatis, K., C. K. Doyle, A. Lykidis, N. Ivanova, M. P. Francino, P. Chain, M. Shin, S. Malfatti, F. Larimer, A. Copeland, J. C. Detter, M. Land, P. M. Richardson, X. J. Yu, D. H. Walker, J. W. McBride, and N. C. Kyrpides. 2006. The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies. J. Bacteriol. 188:4015-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride, J. W., R. E. Corstvet, S. D. Gaunt, C. Boudreaux, T. Guedry, and D. H. Walker. 2003. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 71:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McBride, J. W., J. E. Comer, and D. H. Walker. 2003. Novel immunoreactive glycoprotein orthologs of Ehrlichia spp. Ann. N. Y. Acad. Sci. 990:678-684. [DOI] [PubMed] [Google Scholar]
- 27.McBride, J. W., R. E. Corstvet, E. B. Breitschwerdt, and D. H. Walker. 2001. Immunodiagnosis of Ehrlichia canis infection with recombinant proteins. J. Clin. Microbiol. 39:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride, J. W., C. K. Doyle, X. Zhang, A. M. Cardenas, V. L. Popov, K. A. Nethery, and M. E. Woods. 2006. Identification of a glycosylated Ehrlichia canis 19-kDa major immunoreactive protein with a species-specific serine-rich glycopeptide epitope. Infect. Immun. 75:74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride, J. W., X. Yu, and D. H. Walker. 2000. A conserved, transcriptionally active p28 multigene locus of Ehrlichia canis. Gene 254:245-252. [DOI] [PubMed] [Google Scholar]
- 30.McBride, J. W., X. J. Yu, and D. H. Walker. 1999. Molecular cloning of the gene for a conserved major immunoreactive 28-kilodalton protein of Ehrlichia canis: a potential serodiagnostic antigen. Clin. Diagn. Lab. Immunol. 6:392-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mudaliar, V. S. 1944. Canine rickettsiosis in south India. Indian Vet. J. 20:163. [Google Scholar]
- 32.Ndip, L. M., R. N. Ndip, S. N. Esemu, V. L. Dickmu, E. B. Fokam, D. H. Walker, and J. W. McBride. 2005. Ehrlichial infection in Cameroonian canines by Ehrlichia canis and Ehrlichia ewingii. Vet. Microbiol. 111:59-66. [DOI] [PubMed] [Google Scholar]
- 33.Ndip, L. M., R. N. Ndip, V. E. Ndive, J. A. Awuh, D. H. Walker, and J. W. McBride. 2006. Ehrlichia species in Rhipicephalus sanguineus ticks in Cameroon. Vector Borne Zoonot. Dis. 221-227. [DOI] [PubMed]
- 34.Nethery, K. A., C. K. Doyle, B. L. Elsom, N. K. Herzog, V. L. Popov, and J. W. McBride. 2005. Ankyrin repeat containing immunoreactive 200 kD glycoprotein (gp200) orthologs of Ehrlichia chaffeensis and Ehrlichia canis are translocated to the nuclei of infected monocytes, p. O-60. Abstr. 4th Int. Conf. Rickettsiae Rickettsial Dis., Longrono, Spain.
- 35.Nethery, K. A., C. K. Doyle, X. Zhang, and J. W. McBride. 2007. Ehrlichia canis gp200 contains dominant species-specific antibody epitopes in terminal acidic domains. Infect. Immun. 75:4900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nims, R. M., J. A. Ferguson, J. L. Walker, P. K. Hildebrandt, D. L. Huxsoll, M. J. Reardon, J. E. Varley, G. J. Kolaja, W. T. Watson, E. L. Shroyer, P. A. Elwell, and G. W. Vacura. 1971. Epizootiology of tropical canine pancytopenia in Southeast Asia. J. Am. Vet. Med. Assoc. 158:53-63. [PubMed] [Google Scholar]
- 37.Paddock, C. D., J. W. Sumner, G. M. Shore, D. C. Bartley, R. C. Elie, J. G. McQuade, C. R. Martin, C. S. Goldsmith, and J. E. Childs. 1997. Isolation and characterization of Ehrlichia chaffeensis strains from patients with fatal ehrlichiosis. J. Clin. Microbiol. 35:2496-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez, M., M. Bodor, C. Zhang, Q. Xiong, and Y. Rikihisa. 2006. Human infection with Ehrlichia canis accompanied by clinical signs in Venezuela. Ann. N. Y. Acad. Sci. 1078:110-117. [DOI] [PubMed] [Google Scholar]
- 39.Perez, M., Y. Rikihisa, and B. Wen. 1996. Ehrlichia canis-like agent isolated from a man in Venezuela: antigenic and genetic characterization. J. Clin. Microbiol. 34:2133-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinyoowong, D., S. Jittapalapong, F. Suksawat, R. W. Stich, and A. Thamchaipenet. 2008. Molecular characterization of Thai Ehrlichia canis and Anaplasma platys strains detected in dogs. Infect. Genet. Evol. 8:433-438. [DOI] [PubMed] [Google Scholar]
- 41.Pretorius, A. M., and P. J. Kelly. 1998. Serological survey for antibodies reactive with Ehrlichia canis and E. chaffeensis in dogs from the Bloemfontein area, South Africa. J. S. Afr. Vet. Assoc. 69:126-128. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Vivas, R. I., R. E. Albornoz, and G. M. Bolio. 2005. Ehrlichia canis in dogs in Yucatan, Mexico: seroprevalence, prevalence of infection and associated factors. Vet. Parasitol. 127:75-79. [DOI] [PubMed] [Google Scholar]
- 43.Siarkou, V. I., M. E. Mylonakis, E. Bourtzi-Hatzopoulou, and A. F. Koutinas. 2007. Sequence and phylogenetic analysis of the 16S rRNA gene of Ehrlichia canis strains in dogs with clinical monocytic ehrlichiosis. Vet. Microbiol. 125:304-312. [DOI] [PubMed] [Google Scholar]
- 44.Solano-Gallego, L., M. Trotta, L. Razia, T. Furlanello, and M. Caldin. 2006. Molecular survey of Ehrlichia canis and Anaplasma phagocytophilum from blood of dogs in Italy. Ann. N. Y. Acad. Sci. 1078:515-518. [DOI] [PubMed] [Google Scholar]
- 45.Sumner, J. W., J. E. Childs, and C. D. Paddock. 1999. Molecular cloning and characterization of the Ehrlichia chaffeensis variable-length PCR target: an antigen-expressing gene that exhibits interstrain variation. J. Clin. Microbiol. 37:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unver, A., M. Perez, N. Orellana, H. Huang, and Y. Rikihisa. 2001. Molecular and antigenic comparison of Ehrlichia canis isolates from dogs, ticks, and a human in Venezuela. J. Clin. Microbiol. 39:2788-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Unver, A., Y. Rikihisa, K. Borku, Y. Ozkanlar, and B. Hanedan. 2005. Molecular detection and characterization of Ehrlichia canis from dogs in Turkey. Berl. Munch. Tierarztl. Wochenschr. 118:300-304. [PubMed] [Google Scholar]
- 48.Vinasco, J., O. Li, A. Alvarado, D. Diaz, L. Hoyos, L. Tabachi, K. Sirigireddy, C. Ferguson, and M. H. Moro. 2007. Molecular evidence of a new strain of Ehrlichia canis from South America. J. Clin. Microbiol. 45:2716-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker, J. S., J. D. Rundquist, R. Taylor, B. L. Wilson, M. R. Andrews, J. Barck, A. L. J. Hogge, D. L. Huxsoll, P. K. Hildebrandt, and R. M. Nims. 1970. Clinical and clinicopathologic findings in tropical canine pancytopenia. J. Am. Vet. Med. Assoc. 157:43-55. [PubMed] [Google Scholar]
- 50.Watanabe, M., M. Okuda, M. Tsuji, and H. Inokuma. 2004. Seroepidemiological study of canine ehrlichial infections in Yamaguchi prefecture and surrounding areas of Japan. Vet. Parasitol. 124:101-107. [DOI] [PubMed] [Google Scholar]
- 51.Wilkins, J. H., R. S. Bowden, and G. T. Wilkinson. 1967. A new canine disease syndrome. Vet. Rec. 81:57-58. [DOI] [PubMed] [Google Scholar]
- 52.Yabsley, M. J., S. E. Little, E. J. Sims, V. G. Dugan, D. E. Stallknecht, and W. R. Davidson. 2003. Molecular variation in the variable-length PCR target and 120-kilodalton antigen genes of Ehrlichia chaffeensis from white-tailed deer (Odocoileus virginianus). J. Clin. Microbiol. 41:5202-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu, X. J., J. W. McBride, C. M. Diaz, and D. H. Walker. 2000. Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J. Clin. Microbiol. 38:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]