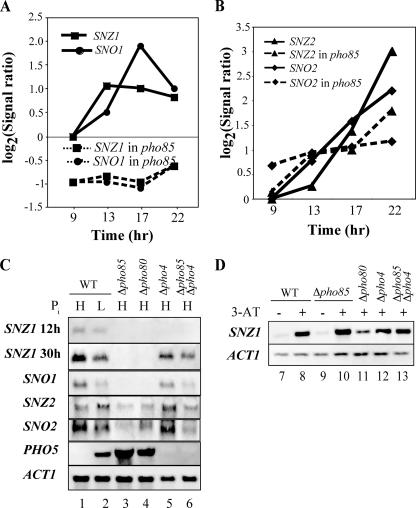
FIG. 1.
Requirement of Pho85 for SNZ/SNO gene expression. (A) Time course of SNZ1/SNO1 expression as determined by GeneChip analysis (based on data from reference 18). The extents of SNZ1 (squares) and SNO1 (circles) expression are shown as log2(signal ratio) values. Solid and broken lines designate expression in the wt and a Δpho85 mutant, respectively. (B) Time course of SNZ2/SNO2 expression by GeneChip analysis (18). SNZ2 (triangles) and SNO2 (diamonds) expression levels in the wt and a Δpho85 mutant are shown as described for panel A. (C) Northern analysis of SNZ1, SNO1, SNZ2, and SNO2 expression in the wt and various pho mutants and under high (H)- or low (L)-Pi conditions. Total RNA was extracted from the yeast cells (designated at the top of the panel) grown for 12 (SNZ1 only) or 30 h and was subjected to Northern analysis as described in Materials and Methods, and then the blot was hybridized with the respective digoxigenin-labeled probe. To analyze PHO5 expression, yeast cells were incubated in high- or low-Pi medium for 5 h before RNA isolation. ACT1 was detected with RNA prepared from a 12-h culture. (D) Effect of 3-AT on SNZ1 expression. Yeast strains, as designated at the top of the panel, were grown to mid-log phase, resuspended in SD medium lacking histidine and with (+) or without (−) 100 mM 3-AT, and then incubated for 1 h before RNA isolation.