FIG. 3.
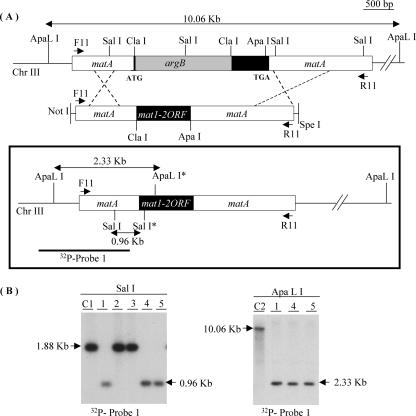
ORF swap strategy. (A) Schematic representation of the WT A. nidulans matA genomic region on Chr III. A. nidulans matA disruption allele and construct used to introduce the A. fumigatus mat1-2 ORF by homologous recombination. The argB selectable marker is incorporated between the Cla1 sites of the A. nidulans matA ORF (black solid box). Position of primers (F11 and R11) used to create the ORF swap construct are indicated. The mat1-2 ORF was cloned between Cla1 and Apa1 as described in Materials and Methods. The flanking regions with A. nidulans matA homology for double-crossover gene replacement are indicated as white boxes. Structure of the A. nidulans matA::mat1-2 genomic region created upon homologous recombination (bold frame) is shown within the box. Enzymes used for Southern blot analysis are indicated. Sal1* and ApaL1* are present exclusively in A. fumigatus mat1-2. The restriction enzymes and probe used to confirm gene replacement are marked. (B) Results of Southern blot analysis. Sal1 digestion differentiates between A. nidulans matA ORF and A. fumigatus mat1-2 ORF. ApaL1 digestion was used to determined the number of integrated mat1-2 copies. C1, control, WT matA in FGSCA26; C2, control, disrupted ΔmatA in UI404.