Abstract
Hfq is a bacterial RNA chaperone involved in the posttranscriptional regulation of many stress-inducible genes via small noncoding RNAs. Here, we show that Hfq is critical for the uropathogenic Escherichia coli (UPEC) isolate UTI89 to effectively colonize the bladder and kidneys in a murine urinary tract infection model system. The disruption of hfq did not affect bacterial adherence to or invasion of host cells but did limit the development of intracellular microcolonies by UTI89 within the terminally differentiated epithelial cells that line the lumen of the bladder. In vitro, the hfq mutant was significantly impaired in its abilities to handle the antibacterial cationic peptide polymyxin B and reactive nitrogen and oxygen radicals and to grow in acidic medium (pH 5.0). Relative to the wild-type strain, the hfq mutant also had a substantially reduced migration rate on motility agar and was less prone to form biofilms. Hfq activities are known to impact the regulation of both the stationary-phase sigma factor RpoS (σS) and the envelope stress response sigma factor RpoE (σE). Although we saw similarities among hfq, rpoS, and rpoE deletion mutants in our assays, the rpoE and hfq mutants were phenotypically the most alike. Cumulatively, our data indicate that Hfq likely affects UPEC virulence-related phenotypes primarily by modulating membrane homeostasis and envelope stress response pathways.
Small noncoding regulatory RNAs (sRNAs) can modulate the translation and stability of specific target mRNAs in prokaryotes and can thereby impact multiple aspects of bacterial cell physiology. In Escherichia coli, more than 60 sRNAs have been conclusively identified, representing 1 to 2% of the number of known protein-encoding genes in this organism (20). Interactions between most sRNA molecules and mRNAs occur through multiple regions of homology of 2 to 8 bp, typically within the 5′ ends of target transcripts (21). In many cases, these RNA-RNA interactions require Hfq, a protein originally identified as a host factor needed for Qβ bacteriophage replication (18, 19). Hfq assembles into homohexameric rings, which are structurally similar to those formed by the Sm and Sm-like proteins that comprise the core of splicing and mRNA degradation complexes in eukaryotic and archaeal cells (21, 52, 56). By binding single-stranded AU-rich regions, Hfq can stabilize sRNA molecules as well as stimulate the formation of sRNA-mRNA pairs. In most cases, these Hfq-mediated interactions have an inhibitory effect on either the translation or the stability of the target mRNA.
A number of sRNA molecules that bind Hfq are key regulatory elements in bacterial stress responses (20). Among these are sRNAs that help control the expression of the sigma factor RpoS (σS), a master regulator of the general stress response in E. coli and many other gram-negative bacteria. RpoS, which is also known as the stationary-phase sigma factor, regulates the expression of numerous genes that promote bacterial survival in the face of various environmental stresses, including nutrient limitation, UV radiation, hyperosmotic shock, temperature extremes, acidic pH, and oxidative stress (23). The sRNA OxyS, which is expressed in response to oxidative stress, represses RpoS translation (59, 60), while the sRNAs DsrA and RprA enhance RpoS expression (31, 47). Very little RpoS is synthesized in an hfq mutant (35, 41), and many of the phenotypic effects observed with an hfq knockout have been attributed to defects in RpoS expression (36).
Recent work has revealed that the deletion of hfq also impacts the envelope stress response sigma factor, RpoE (σE) (25, 46, 54, 13, 16). RpoE is activated in response to extracytoplasmic stresses, like heat shock or misfolded outer membrane proteins (OMPs), and regulates the expression of about 100 genes. These include the sRNAs MicA, which inhibits OmpA expression, and RybB, which inhibits both OmpC and OmpW expression. Together with other RpoE regulon members, these sRNAs in association with Hfq help maintain envelope integrity by coordinating the expression of OMPs and other bacterial envelope components (5, 25, 54, 55). Interestingly, RybB also represses RpoE translation, creating an autoregulatory loop (54). The deletion of hfq causes strong activation of RpoE, probably due to diminished RybB activity, coupled with misregulated OMP expression and increased envelope stress. This stress, in turn, likely stimulates the activation of the periplasmic protease DegS and the subsequent degradation of the RpoE sequestration factor RseA (16).
Stress tolerance is central to the ability of many bacterial pathogens to successfully colonize hostile host environments. Considering the roles of Hfq and sRNAs as key regulators of stress response pathways in laboratory E. coli K-12 strains (20), we were interested in understanding how Hfq might contribute to the virulence of uropathogenic E. coli (UPEC) bacteria. These bacteria are the primary cause of urinary tract infections (UTIs), including both cystitis (bladder infection) and pyelonephritis (kidney infection) (17). UTIs are among the most common infections, representing an enormous financial and health burden worldwide (17, 29). The successful colonization of the urinary tract requires that UPEC overcome a barrage of innate host defenses, including the shear flow of urine, the synthesis of soluble and tissue-associated antibacterial molecules, the influx of neutrophils, the exfoliation and clearance of infected host epithelial cells, and the generation of reactive nitrogen species (RNS) and reactive oxygen species (ROS) (6, 32, 39, 42). To counter these defenses, UPEC encodes numerous virulence factors, including various adhesins, toxins, iron chelators, capsule-forming polysaccharides, and flagella (6, 26, 42). The ability of UPEC to invade host epithelial cells, multiply intracellularly, and form biofilms also enhances UPEC virulence and persistence within the urinary tract (6, 37, 38, 50).
Here, we employ a mouse UTI model system to show that Hfq is critical to the ability of UPEC to effectively colonize and persist within the urinary tract. Using in vitro assays, we demonstrate that Hfq affects a number of virulence-related UPEC phenotypes, including biofilm formation, motility, and resistance to RNS, ROS, and the antimicrobial peptide polymyxin B. In addition, we show in comparative analyses that hfq, rpoS, and rpoE UPEC mutants have partly overlapping, yet distinct, phenotypes.
MATERIALS AND METHODS
Strains and plasmids.
Bacterial strains used in this study are listed in Table 1. Mutants were constructed with the human cystitis isolate UTI89 (9, 38) by using the lambda Red recombinase method as described previously (11, 40). Antibiotic resistance cassettes were amplified from Salmonella strain TT23216 or TT23691 chromosomal templates by PCR using primers listed in Table S1 in the supplemental material. TT23216 and TT23691 (strains containing either a chloramphenicol or a kanamycin resistance cassette flanked by “universal ends” for use in generating knockouts by lambda Red-mediated recombination) were kindly provided by John Roth (University of California, Davis). All primers were designed with overhangs specific for the first 40 bp within or surrounding the 5′ and 3′ ends of the target UTI89 genes. PCR products were introduced by electroporation into UTI89 carrying pKM208, which encodes IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible lambda Red recombinase. Knockouts were verified by PCR using flanking primers specific for each targeted gene.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| UTI89 | Wild-type cystitis isolate | 38 |
| MM835 | UTI89 Δhfq::Clmr | This study |
| MM793 | UTI89 ΔrpoS::Kanr | This study |
| MM836 | UTI89 Δ(rpoE-rseABC)::Clmr | This study |
| MM837 | UTI89 ΔhflX::Clmr | This study |
| ES707 | UTI89 with pKM208 | This study |
| MM807 | UTI89 with pACYC177 | This study |
| MM808 | UTI89 Δhfq::Clmr with pHfq | This study |
| MM810 | UTI89 Δhfq::Clmr with pACYC177 | This study |
| MM803 | UTI89 Δhfq::Clmr with pKM208 | This study |
| MM905 | UTI89 Δ(rpoE-rseABC)::Clmr with pACYC177 | This study |
| MM906 | UTI89 Δ(rpoE-rseABC)::Clmr with pJLJ41 | This study |
| MM904 | UTI89 ΔrpoS::Kanr with pACYC177 | This study |
| MM 903 | UTI89 ΔrpoS::Kanr with pRpoS4 | This study |
| Top10 | Ultracompetent strain | Invitrogen |
| Salmonella | ||
| TT23216 | Strain with Clmr cassette flanked by universal primer sites | John Roth |
| TT23691 | Strain with Kanr cassette flanked by universal primer sites | John Roth |
| Plasmids | ||
| pKM208 | IPTG-inducible lamda Red recombinase expression plasmid; Ampr | 40 |
| pCR2.1 | High-copy-no. cloning vector | Invitrogen |
| pACYC177 | Low-copy-no. vector containing Ampr and Kanr cassettes | New England Biolabs |
| pHfq | hfq cloned from UTI89 with native promoter ligated into pACYC177 backbone; contains Ampr cassette | This study |
| pRpoS4 | rpoS cloned from UTI89 with native promoter ligated into pACYC177 backbone; contains Ampr cassette | This study |
| pJLJ41 | rpoE-rseABC from UTI89 with native promoter ligated into pACYC177 backbone; contains Kanr cassette | This study |
Primers used for cloning hfq, rpoS, and rpoE-rseABC from UTI89 are listed in Table S1 in the supplemental material. The hfq gene, along with 300 bp of upstream sequence, was amplified using whole-colony PCR with primers P112 and P113. The PCR product was ligated into pCR2.1 (Invitrogen), sequenced, and subsequently subcloned into the low-copy-number plasmid pACYC177 (New England Biolabs) by using BamHI and XhoI restriction sites to create pHfq. The rpoS gene plus 200 bp of upstream sequence was similarly cloned using primers P180 and P181, creating pRpoS4. The rpoE and rseABC genes, in addition to 350 bp of upstream and 100 bp of downstream sequences, were cloned using primers F_rpoE and R_rseC. The PCR product was digested and ligated directly into pACYC177 using BamHI and PstI restriction sites to make pJLJ41. In complementation experiments, pACYC177 served as an empty-vector control.
Mouse infections.
Cultures of UTI89 and the hfq knockout mutant (UTI89 Δhfq) from freezer stocks were grown in 20 ml of Luria-Bertani (LB) broth for 48 h at 37°C without shaking. Just prior to infection, bacteria from these cultures were pelleted and resuspended in phosphate-buffered saline (PBS). Seven-week-old female CBA/J mice (Jackson Laboratory) were briefly anesthetized with isoflurane and inoculated transurethrally with 50 μl of the bacterial suspension (approximately 108 bacteria, as determined by plating) as described previously (37). At days 1, 3, and 5 postinoculation, the mice were sacrificed and the bladder and left kidney of each animal were harvested aseptically, weighed, and homogenized in 1 ml of PBS containing 0.025% Triton X-100. Homogenates were serially diluted and plated onto LB agar plates to determine bacterial titers. Mouse experiments were repeated twice, with similar results.
The formation of intracellular bacterial communities (IBCs) by UTI89 and the Δhfq mutant was quantified as reported previously (28). Bladders from 7-week-old female CBA/J or C3H/HeN mice (Harlan) were recovered 6 h postinoculation with equal numbers of CFU of either wild-type UTI89 or the Δhfq mutant, halved, splayed, and fixed in 10% neutral buffered formalin for 30 min. Bladders were rinsed twice with wash buffer (0.01 M MgCl2, 0.02% octylphenoxypolyethoxyethanol [Igepal], and 0.01% sodium deoxycholate in PBS) and incubated overnight at 4°C in lacZ stain buffer (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 1 mg of X-Gal [5-bromo-4-chloro-3-indolylphosphate]/ml in wash buffer). Stained bladder halves were then washed in PBS, mounted under coverslips, and viewed using bright-field optics under an Olympus BX51 microscope. All mouse experiments were performed under accredited conditions using Institutional Animal Care and Use Committee-approved protocols.
Growth assays.
Cultures of UTI89 and its derivatives were grown overnight at 37°C in 5 ml of LB broth, 100 mM morpholineethanesulfonic acid (MES)-buffered LB (LB-MES; pH 5.0), or M9 minimal medium (6 g of Na2HPO4/liter, 3 g of KH2PO4/liter, 1 g of NH4Cl/liter, 0.5 g of NaCl/liter, 1 mM MgSO4, 0.1 mM CaCl2, 0.1% glucose, 0.0025% nicotinic acid, and 16.5 μg of thiamine/ml in H2O) in loosely capped 20- by 150-mm borosilicate glass tubes with shaking (225 rpm, with the tubes tilted at a 30° angle). Each culture was diluted to an A600 of ∼1.0 and then subcultured at 1:100 in LB, LB-MES, or M9 medium. Growth in LB with or without 1 mM methyl viologen (MV) and in LB-MES with or without 1 mM sodium nitrite (Sigma-Aldrich) was assessed. All growth curves were generated from quadruplicate 200-μl cultures in 100-well honeycomb plates at 37°C with shaking by using a Bioscreen C instrument (Growth Curves USA). Overnight cultures of strains carrying pACYC177, pHfq, pRpoS4, or pJLJ41 for complementation experiments were grown in the presence of 100 μg of ampicillin/ml or 50 μg of kanamycin/ml to maintain the plasmids, but the antibiotics were not added to media used in the subsequent assays.
Biofilm assays.
In vitro biofilm formation assays were performed as described previously (34). Briefly, 5-ml cultures of UTI89 and its derivatives were first grown overnight with shaking at 37°C in M9 medium. These bacteria were diluted 1:100 in M9 medium, and quadruplicate 100-μl samples in 96-well pinchbar flat-bottomed polystyrene microtiter plates with lids (NUNC) were incubated without shaking for 48 h at 37°C. Planktonic bacteria were then removed by inverting and shaking the plates vigorously and washing them twice with double-distilled water. Crystal violet (150 μl of a 0.1% solution in water; Sigma-Aldrich) was added to each well, and the plates were incubated at room temperature for an additional 10 min. After the removal of the crystal violet, the wells were washed twice with double-distilled water and air dried at room temperature. Dimethyl sulfoxide (200 μl; Sigma-Aldrich) was added to each well, and the plates were again shaken vigorously for 15 min at room temperature. A 150-μl aliquot from each well was transferred onto a fresh microtiter plate, and A562 readings were taken using a Synergy HT multidetection microplate reader (Biotek Instruments, Inc.).
Agglutination, cell association, and invasion assays.
The ability of UTI89 and its derivatives to agglutinate Saccharomyces cerevisiae cells was qualitatively determined by mixing 20 μl of each bacterial strain (from overnight static cultures) with 200 μl of a 1% suspension of baker's yeast in PBS on glass slides. Hemagglutination assays were carried out using guinea pig red blood cells (Colorado Serum Company) according to established protocols (48). Bacterial host cell association and invasion assays were performed using human bladder epithelial cells (designated 5637 cells) and the A498 human kidney cell line (American Type Culture Collection) as described previously (14).
Motility assays.
Prewarmed motility agar plates, containing 0.2% agar (EMD Chemicals) in LB broth, were inoculated (on the surface) with 1 μl of bacteria from overnight cultures that had been grown with shaking and subsequently diluted to an A600 of ∼1.0. Plates were incubated at 37°C, and bacterial spreading (swarming) was measured at 2-h intervals and photographed using a Nikon D80 digital camera.
Polymyxin B sensitivity assays.
Bacterial cultures, grown with shaking overnight at 37°C, were diluted in LB broth to an A600 of ∼1.0 and subcultured at 1:100 in 5 ml of LB broth containing 0, 1, or 5 μg of polymyxin B (Sigma-Aldrich)/ml. These cultures were incubated at 37°C with shaking at 225 rpm for 1.5 h, and bacterial titers were then determined by plating serial dilutions of each sample.
LPS profiling.
Lipopolysaccharide (LPS) profiling was performed as described previously (3, 24). Briefly, bacterial cultures were grown to stationary phase in LB broth at 37°C and normalized to an A600 of 1.0. Bacteria from 1 ml of each sample were pelleted and resuspended in 250 μl of water prior to the addition of 250 μl of bacterial lysis buffer (1% sodium dodecyl sulfate, 50 mM Tris-Cl [pH 7.0], 10 mM EDTA). After boiling for 5 min, samples were incubated with proteinase K (1.5 mg/ml; Sigma-Aldrich) for 3 h at 37°C. LPS extracts were subsequently resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 12.5% polyacrylamide gels, which were then stained using the SilverSnap stain kit II (Pierce).
Statistics.
Mann-Whitney U and two-tailed unpaired t tests were performed using Prism 5.01 software (GraphPad Software). P values of less than 0.05 were considered significant.
RESULTS
The disruption of hfq attenuates UPEC colonization of the urinary tract.
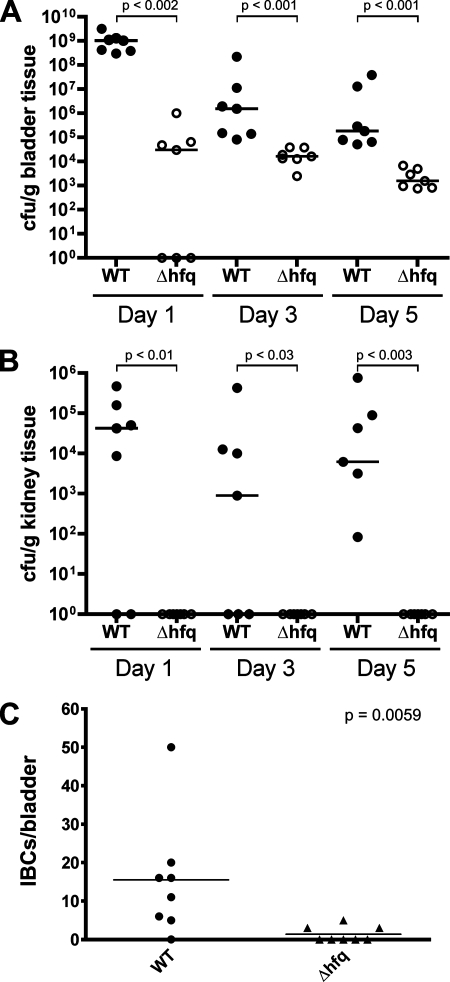
To assess the role of Hfq in cystitis and pyelonephritis caused by UPEC, adult female CBA/J mice were inoculated via transurethral catheterization with a wild-type UPEC cystitis isolate, UTI89, or the hfq knockout mutant UTI89 Δhfq. At 1, 3, and 5 days postinoculation, the bladders and kidneys were collected and homogenized and bacterial titers were determined by serial dilution and plating of the tissue homogenates. As shown in Fig. 1, the hfq mutant was severely impaired at all time points in its abilities to colonize and persist within both the kidneys and the bladder relative to the wild-type UTI89 strain. However, in vitro, UTI89 Δhfq grew similarly to the wild-type strain in both LB broth and M9 minimal medium (data not shown). Together, these results indicate that Hfq contributes significantly to the fitness of UPEC within the urinary tract.
FIG. 1.
Hfq is required for effective UPEC colonization of the urinary tract. (A and B) Adult female CBA/J mice were infected with 108 CFU of UTI89 or UTI89 Δhfq via transurethral catheterization. Bacterial titers in bladder (A) and kidney (B) homogenates were determined at the indicated times postinoculation. Horizontal bars indicate median values for each group. WT, wild type. (C) The graph shows total numbers of IBCs per bladder at 6 h postinoculation of CBA/J mice with either UTI89 or UTI89 Δhfq. Bars denote mean values. The indicated P values were determined using the Mann-Whitney U test (n = 7 or 8 mice per group).
In vitro assays revealed no defects in the ability of UTI89 Δhfq to interact with host cells: the hfq mutant agglutinated yeast and red blood cells normally and was able to bind to and invade both bladder and kidney epithelial cells at wild-type levels in cell culture-based assays (data not shown). Within the superficial epithelial cells that line the luminal surface of the bladder, UTI89 and other UPEC isolates are able to multiply rapidly, forming large cytosolic inclusions referred to as pods, or IBCs (2, 27, 15, 38). UTI89 bacteria within IBCs naturally express LacZ, enabling these inclusions to be visualized and enumerated in whole-mount bladder preparations when stained using X-Gal (28). By 6 h post-transurethral inoculation of adult female CBA/J mice, we detected at least a few IBCs in most of the UTI89-infected bladders but no IBCs were detected in the majority of bladders infected with UTI89 Δhfq (Fig. 1C). Similar results were obtained using C3H/HeN mice. Importantly, the hfq mutant was able to express wild-type levels of beta-galactosidase (LacZ) activity, and no X-Gal staining of mock-infected bladders was detected (data not shown). These data, coupled with our observations that UTI89 Δhfq bound to and invaded host cells normally, indicate a defect in the ability of the hfq mutant to multiply intracellularly.
Hfq affects UPEC growth at low pH and resistance to RNS and ROS.
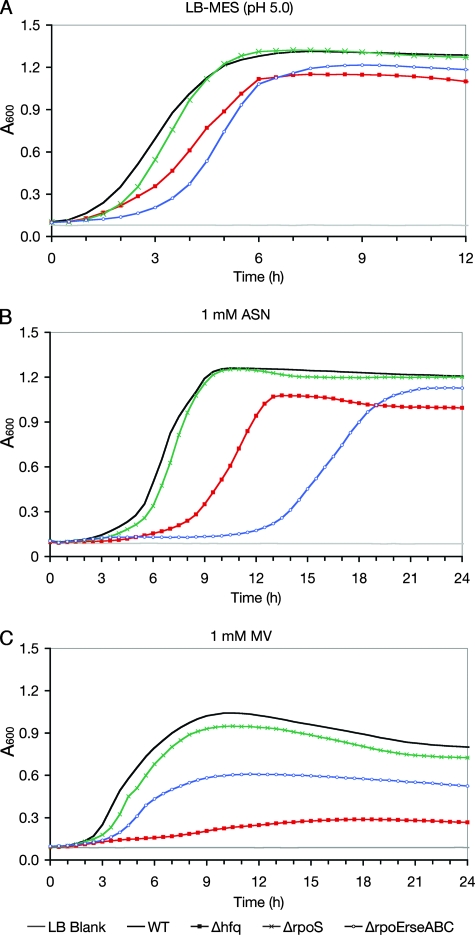
Within bladder epithelial cells, UPEC is initially trafficked into acidic, late-endosome-like compartments where bacterial replication can be initiated prior to the development of IBCs (15). UPEC may encounter similarly acidic environments within urine during the acute phase of a UTI. In addition, UPEC must deal with high levels of ROS and RNS that are generated within the urinary tract during infection. UPEC isolates like UTI89 often have much greater resistance to these radicals than laboratory E. coli K-12 strains (7, 42, 53). A role for Hfq during bacterial growth under acidic conditions was assessed using LB-MES (pH 5.0), while the contribution of Hfq to UPEC RNS and ROS resistance was tested using acidified sodium nitrite (ASN) and MV, respectively. When added to LB-MES (pH 5.0), sodium nitrite is converted into nitrous acid, which spontaneously decomposes to form NO and other RNS (57). MV, on the other hand, generates superoxide radicals (22). We found that the growth of UTI89 Δhfq, which grew normally in LB broth (pH 7.0), consistently lagged behind that of the wild-type strain by more than an hour when the strains were grown in LB-MES (pH 5.0) (Fig. 2A). The relative growth of UTI89 Δhfq in 1 mM ASN was even more severely impaired (Fig. 2B), and in the presence of 1 mM MV, the hfq mutant barely grew at all (Fig. 2C). Controls for these and other in vitro assays described in the following sections included UTI89 Δhfq strains complemented with pHfq (a low-copy-number plasmid for the expression of hfq from its native promoter) or the empty vector pACYC177. Possible polar effects resulting from hfq deletion in UTI89 were controlled for by disrupting hflX, which is immediately downstream from and in frame with hfq. The hflX gene encodes a GTP-binding protein of unknown function. In all assays, the hflX mutant and UTI89 Δhfq complemented with pHfq behaved like the wild type (data not shown). Cumulatively, these data indicate that the Hfq RNA chaperone enhances bacterial growth under acidic conditions and that Hfq has an especially important role in UPEC resistance to both ROS and RNS, possibly via indirect effects on the expression of stress-responsive genes.
FIG. 2.
UPEC resistance to low pH, RNS, and ROS is differentially affected by Hfq, RpoS, and RpoE. Overnight cultures of wild-type UTI89 (WT), UTI89 Δhfq, UTI89 ΔrpoS, and UTI89 Δ(rpoE-rseABC) were diluted to an A600 of 1.0 and subcultured at 1:100 in LB-MES (pH 5.0) (A), LB-MES-1 mM ASN (B), or LB broth-1 mM MV (C). Cultures were grown in plate format, and absorbance measurements were obtained using a Bioscreen C instrument (Growth Curves USA). Each growth curve represents the means of results for quadruplicate samples, and each experiment was repeated three or more times, with similar results.
Effects of Hfq on UPEC motility.
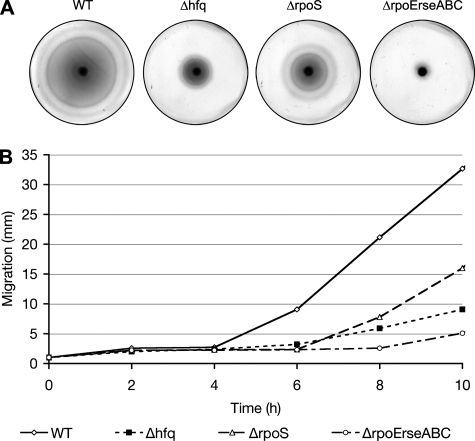
The disruption of hfq impairs the motility of at least two pathogens, Salmonella enterica serovar Typhimurium and Pseudomonas aeruginosa (46, 49). Within the urinary tract, motility gives UPEC a survival advantage, enhancing bacterial colonization and persistence (58, 30). In consideration of these data, we wished to determine if Hfq affected the motility of UTI89. By light microscopy, we observed that both wild-type UTI89 and UTI89 Δhfq LB broth cultures contained numerous motile microbes, indicating that Hfq is not an absolute requirement for UTI89 motility. However, on motility agar plates, UTI89 Δhfq showed greatly reduced outward migration (swarming) in comparison with the wild-type parent strain (Fig. 3). This motility defect was eliminated by complementation with pHfq but not with the empty-vector control pACYC177 and was not observed with UTI89 ΔhflX (data not shown). All of the strains tested eventually spread across the agar plates, forming concentric rings, like wild-type UTI89, that were characteristic of motile, chemotactic bacteria. These results confirm our microscopic observations that hfq is not required for UTI89 motility while also suggesting a role for Hfq as a modulator of UPEC motility rates and/or chemotaxis.
FIG. 3.
Effects of hfq, rpoS, and rpoE-rseABC disruption on UPEC motility. (A) Images show the spread of UTI89 (the wild type [WT]) and its derivatives at 37°C 8 h after inoculation onto motility agar plates. (B) The change over time in the diameter (in millimeters) of the area covered by each bacterial strain as it spread across the motility plate is represented in the graph. These experiments were repeated three times, with similar results.
Hfq functions in biofilm formation.
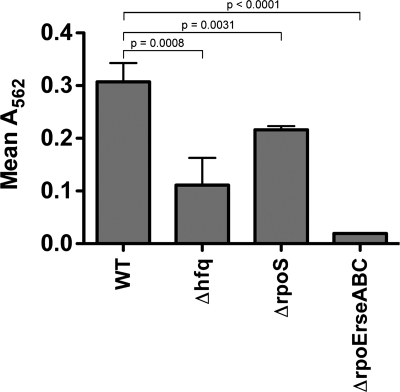
Pathogenic E. coli strains and other bacteria are more prone to form biofilms when nutrient levels are suboptimal, indicating that biofilm formation may act as an adaptation to nutrient-poor environments, as found within the urinary tract (12, 51). Recently, it was noted that UPEC strains that cause relapsing UTIs in women are generally better able to form biofilms than other UPEC isolates in microtiter plate-based assays, suggesting a role for biofilm formation in the establishment and persistence of UPEC within the host (50). The involvement of Hfq in the development of biofilms by UPEC was tested by growing UTI89 and its mutant derivatives at 37°C in M9 minimal medium in 96-well polystyrene microtiter plates. After 48 h, all planktonic bacteria were removed and any remaining bacterial biofilms were stained and quantified using crystal violet. In these assays, UTI89 Δhfq showed about a threefold reduction in biofilm formation relative to the wild-type strain (Fig. 4). Biofilm formation was restored to wild-type levels by complementation with pHfq, and no problems with UTI89 ΔhflX were observed (data not shown). Notably, the biofilm deficiency seen with the hfq mutant was not attributable to growth defects in M9 medium or to any inherent inability of UTI89 Δhfq to retain crystal violet.
FIG. 4.
Effects of hfq, rpoS, and rpoE-rseABC disruption on biofilm formation by UTI89. The wild-type (WT) UTI89 strain and its derivatives were grown in static M9 medium on flat-bottomed microtiter plates at 37°C for 48 h. Added crystal violet that was retained within wells by bacterial biofilms was eluted into dimethyl sulfoxide. The optical density (A562) of this solution correlated with the level of biofilm formation. The graph shows the means for each sample set ± standard deviations, with indicated P values determined by two-tailed unpaired t tests. Of note, planktonic wild-type and mutant bacteria stain equally well with crystal violet in Gram staining assays.
Hfq is required for UPEC resistance to polymyxin B.
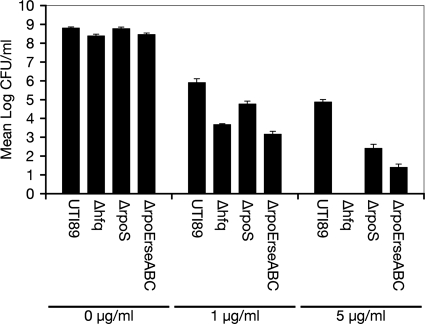
Antimicrobial peptides such as defensins and cathelicidins are important components of the host defense against uropathogenic bacteria (61). These cationic peptides can associate with bacterial membranes, perturbing the integrity of the envelope and potentially disturbing other bacterial components. The capacity of Hfq to influence RpoS and RpoE envelope stress response pathways led us to hypothesize that an hfq deletion mutant might be compromised in its ability to deal with the membrane-disrupting activities of antimicrobial cationic peptides. To test this possibility, we employed the cationic peptide polymyxin B, which has been used clinically as a bactericidal antibiotic. Equal numbers of CFU of UTI89 and its derivatives were incubated for 90 min with shaking in LB broth alone or LB broth with polymyxin B added to a final concentration of 1 or 5 μg/ml. The strains grew to similar titers in the absence of polymyxin B, reaching about 109 CFU/ml (Fig. 5). However, relative to the wild-type strain, UTI89 Δhfq showed pronounced sensitivity to 1- and 5-μg/ml concentrations of polymyxin B. Complementation with pHfq restored the growth of UTI89 Δhfq to wild-type levels, while UTI89 ΔhflX behaved like the wild type in these assays (data not shown). These results indicate a critical role for Hfq in the resistance of UPEC to antimicrobial peptides.
FIG. 5.
UTI89 Δhfq is hypersensitive to the cationic peptide polymyxin B. Equal numbers of CFU of UTI89 and its mutant derivatives were subcultured in LB broth with or without 1.0 or 5.0 μg of polymyxin B/ml, as indicated. After shaking at 37°C for 1.5 h, bacterial titers (in CFU per milliliter) in triplicate samples were determined by plating serial dilutions. The graph shows the means for each sample set ± standard deviations. These experiments were repeated three times, with similar results.
Phenotypic overlap among hfq, rpoS, and rpoE mutants.
In light of previous observations indicating that many of the phenotypic effects observed with an hfq knockout in E. coli K-12 may be attributed to defects in RpoS expression (35), we disrupted the rpoS gene in UTI89 to create UTI89 ΔrpoS for comparison with UTI89 Δhfq. Interestingly, in our assays we found that UTI89 ΔrpoS behaved remarkably like the wild-type UPEC strain in its abilities to grow in acidic medium (pH 5.0) and to handle RNS and ROS (Fig. 2). Although worse off than the wild-type strain in other assays, UTI89 ΔrpoS was still more motile and more resistant to polymyxin B than UTI89 Δhfq (Fig. 3 and 5). The rpoS mutant, which grew normally in M9 and LB media, was also diminished in its capacity to form biofilms in vitro but was still somewhat better at forming biofilms than UTI89 Δhfq (Fig. 4). The latter result correlates with earlier work implicating RpoS as an important factor during biofilm development by E. coli (1). The complementation of UTI89 ΔrpoS with the plasmid pRpoS4, encoding RpoS under the control of its native promoter, restored the wild-type phenotype in all assays (data not shown). Together, these data indicate that the aberrant phenotypes associated with UTI89 Δhfq are not solely a consequence of diminished RpoS expression.
Because Hfq can affect RpoE- as well as RpoS-mediated stress responses, phenotypic comparisons between UTI89 Δhfq and an rpoE mutant, UTI89 Δ(rpoE-rseABC), were also made. The rpoE mutant strain was constructed so that the rseABC genes, which are adjacent to rpoE and regulate RpoE activation, were also disrupted. In contrast to results with laboratory E. coli K-12 strains (8), the deletion of rpoE in UTI89 was not lethal. Using in vitro assays, we found that UTI89 Δ(rpoE-rseABC) had substantial defects in motility, biofilm formation, and growth in low-pH medium, as well as increased sensitivity to RNS, ROS, and polymyxin B (Fig. 2 to 5). In control assays, the rpoE-rseABC mutant grew normally in both LB and M9 media, and complementation with a plasmid carrying rpoE-rseABC under the control of the native promoter restored the wild-type phenotype (data not shown). In total, these results show that UTI89 Δ(rpoE-rseABC), rather than UTI89 ΔrpoS, was phenotypically the most similar to the hfq mutant, indicating a possible link between Hfq effects on the bacterial envelope and the multiple phenotypic defects we found to be associated with UTI89 Δhfq.
DISCUSSION
The RNA chaperone Hfq contributes to the fitness and virulence of several pathogens, including the gram-negative bacteria Brucella abortus (45), P. aeruginosa (49), Vibrio cholerae (13), Legionella pneumophila (33), and Salmonella serovar Typhimurium (46), as well as the gram-positive organism Listeria monocytogenes (10). Although direct comparisons have not been made, it appears that the spectrum and severity of mutant phenotypes observed upon the deletion of hfq can vary significantly among the different pathogens so far analyzed. For example, both Salmonella serovar Typhimurium and B. abortus hfq mutants are unable to multiply well within host cells, while hfq is dispensable for the normal intracellular growth of L. monocytogenes (10, 45, 46). Here, we have shown that hfq is required for UPEC to effectively colonize and persist within the urinary tract. As seen with Salmonella serovar Typhimurium and B. abortus (45, 46), the deletion of hfq appears to attenuate the intracellular growth of UPEC, interfering with the ability of this pathogen to form IBCs. The disruption of hfq also diminished the capacity of UPEC to tolerate RNS, ROS, and the antibacterial cationic peptide polymyxin B and to grow in acidic medium (pH 5.0). Furthermore, the hfq mutant had reduced motility and chemotaxis and was significantly impaired in its ability to form biofilms, a trait that has previously been associated with decreased bacterial persistence within the urinary tract (50).
During the course of an infection, Hfq likely synergizes with multiple signaling pathways and sigma factors, including RpoE and RpoS, in order to facilitate the resistance and adaptation of UPEC to hostile host environments. Accordingly, in our assays we saw significant phenotypic overlap among the hfq, rpoS, and rpoE-rseABC mutants, with UTI89 Δhfq and UTI89 Δ(rpoE-rseABC) being the most alike. In a laboratory E. coli K-12 strain, as well as in Salmonella serovar Typhimurium and V. cholerae, the deletion of hfq has been shown to induce RpoE activation (13, 16, 25, 46, 54). Strikingly, in V. cholerae, enhanced RpoE activation accounts for the effects on nearly half the genes that are upregulated in an hfq mutant (13). RpoE activation in the absence of Hfq likely occurs in E. coli due to increased envelope stress resulting from the aberrant expression of OMPs and other factors, as well as the inability of the Hfq-dependent sRNA RhyB to effectively inhibit RpoE translation (54). Enhanced RpoE activation probably helps ameliorate some of the deleterious effects of hfq disruption, but the overstimulation of RpoE may also contribute to these problems by further disturbing the balance of factors involved in membrane repair and maintenance. This possibility has not yet been rigorously tested with UPEC, although the effects of RpoE inactivation have been partially explored. In particular, the deletion of DegS, a protease that indirectly activates RpoE by freeing it from RseA repression, was found to attenuate UPEC virulence in a mouse UTI model (43). Similarly, the disruption of degP, skp, or surA, all of which are members of the RpoE regulon, also decreases UPEC virulence in mice (28, 44). Interestingly, it has been shown previously for Salmonella serovar Typhimurium that RpoE can positively regulate hfq expression indirectly via the transcriptional activation of another alternate sigma factor, RpoH (σH) (4). A similar relationship between RpoE activation and enhanced Hfq expression in UPEC may account for some of the phenotypic overlap seen between the rpoE-rseABC and hfq knockout mutants in our assays.
Cumulatively, our results indicate a functional link between Hfq, envelope stress, membrane homeostasis, and the virulence potential of UPEC. This connection is further supported by observations that the disruption of hfq, but not rpoS or rpoE, causes marked alterations in the LPS profile of UTI89 (see Fig. S1 in the supplemental material). This effect in turn may significantly influence the overall fitness of UPEC and susceptibility to antibacterial cationic peptides and other stresses encountered within the host. The capacity of Hfq to affect LPS biogenesis and multiple virulence-related phenotypes in UPEC probably reflects the ability of this chaperone to interact with a wide range of different regulatory RNAs, many of which remain to be operationally defined.
Supplementary Material
Acknowledgments
This study was funded by NIH grants DK068585 and DK069526. R.R.K. and D.S.E. were supported by NIH microbial pathogenesis training grant T32 AI055434, and E.S.S. was supported by NIH postdoctoral fellowship DK070507. K.D.-P. contributed to this work as part of the BioURP Research Experience for Undergraduates Program funded by the National Science Foundation.
Editor: F. C. Fang
Footnotes
Published ahead of print on 5 May 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Adams, J. L., and R. J. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ Microbiol. 654285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, G. G., J. J. Palermo, J. D. Schilling, R. Roth, J. Heuser, and S. J. Hultgren. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301105-107. [DOI] [PubMed] [Google Scholar]
- 3.Aucken, H. M., and T. L. Pitt. 1993. Lipopolysaccharide profile typing as a technique for comparative typing of gram-negative bacteria. J. Clin. Microbiol. 311286-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bang, I. S., J. G. Frye, M. McClelland, J. Velayudhan, and F. C. Fang. 2005. Alternative sigma factor interactions in Salmonella: óE and óH promote antioxidant defences by enhancing óS levels. Mol. Microbiol. 56811-823. [DOI] [PubMed] [Google Scholar]
- 5.Bossi, L., and N. Figueroa-Bossi. 2007. A small RNA downregulates LamB maltoporin in Salmonella. Mol. Microbiol. 65799-810. [DOI] [PubMed] [Google Scholar]
- 6.Bower, J. M., D. S. Eto, and M. A. Mulvey. 2005. Covert operations of uropathogenic Escherichia coli within the urinary tract. Traffic 618-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bower, J. M., and M. A. Mulvey. 2006. Polyamine-mediated resistance of uropathogenic Escherichia coli to nitrosative stress. J. Bacteriol. 188928-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Button, J. E., T. J. Silhavy, and N. Ruiz. 2007. A suppressor of cell death caused by the loss of σE downregulates extracytoplasmic stress responses and outer membrane vesicle production in Escherichia coli. J. Bacteriol. 1891523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, S. L., C. S. Hung, J. Xu, C. S. Reigstad, V. Magrini, A. Sabo, D. Blasiar, T. Bieri, R. R. Meyer, P. Ozersky, J. R. Armstrong, R. S. Fulton, J. P. Latreille, J. Spieth, T. M. Hooton, E. R. Mardis, S. J. Hultgren, and J. I. Gordon. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA 1035977-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christiansen, J. K., M. H. Larsen, H. Ingmer, L. Sogaard-Andersen, and B. H. Kallipolitis. 2004. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 1863355-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewanti, R., and A. C. Wong. 1995. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int. J. Food Microbiol. 26147-164. [DOI] [PubMed] [Google Scholar]
- 13.Ding, Y., B. M. Davis, and M. K. Waldor. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 53345-354. [DOI] [PubMed] [Google Scholar]
- 14.Eto, D. S., T. A. Jones, J. L. Sundsbak, and M. A. Mulvey. 2007. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog. 3e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eto, D. S., J. L. Sundsbak, and M. A. Mulvey. 2006. Actin-gated intracellular growth and resurgence of uropathogenic Escherichia coli. Cell. Microbiol. 8704-717. [DOI] [PubMed] [Google Scholar]
- 16.Figueroa-Bossi, N., S. Lemire, D. Maloriol, R. Balbontin, J. Casadesus, and L. Bossi. 2006. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 62838-852. [DOI] [PubMed] [Google Scholar]
- 17.Foxman, B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A)5S-13S. [DOI] [PubMed] [Google Scholar]
- 18.Franze de Fernandez, M. T., L. Eoyang, and J. T. August. 1968. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 219588-590. [DOI] [PubMed] [Google Scholar]
- 19.Franze de Fernandez, M. T., W. S. Hayward, and J. T. August. 1972. Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 247824-831. [PubMed] [Google Scholar]
- 20.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21399-404. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58303-328. [DOI] [PubMed] [Google Scholar]
- 22.Hassan, H. M., and I. Fridovich. 1978. Superoxide radical and the oxygen enhancement of the toxicity of paraquat in Escherichia coli. J. Biol. Chem. 2538143-8148. [PubMed] [Google Scholar]
- 23.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol Rev. 66373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansen, J., A. A. Rasmussen, M. Overgaard, and P. Valentin-Hansen. 2006. Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins. J. Mol. Biol. 3641-8. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 480-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Justice, S. S., C. Hung, J. A. Theriot, D. A. Fletcher, G. G. Anderson, M. J. Footer, and S. J. Hultgren. 2004. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA 1011333-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Justice, S. S., S. R. Lauer, S. J. Hultgren, and D. A. Hunstad. 2006. Maturation of intracellular Escherichia coli communities requires SurA. Infect. Immun. 744793-4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kucheria, R., P. Dasgupta, S. H. Sacks, M. S. Khan, and N. S. Sheerin. 2005. Urinary tract infections: new insights into a common problem. Postgrad. Med. J. 8183-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, M. C., C. J. Alteri, S. N. Smith, and H. L. Mobley. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. USA 10416669-16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Majdalani, N., S. Chen, J. Murrow, K. St. John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 391382-1394. [DOI] [PubMed] [Google Scholar]
- 32.Mak, R. H., and H. J. Kuo. 2006. Pathogenesis of urinary tract infection: an update. Curr. Opin. Pediatr. 18148-152. [DOI] [PubMed] [Google Scholar]
- 33.McNealy, T. L., V. Forsbach-Birk, C. Shi, and R. Marre. 2005. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J. Bacteriol. 1871527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merritt, T., D. Kadouri, and G. O'Toole. 2005. Growing and analyzing static biofilms, p. 1B.1.1-1B.1.7. In R. Coico, T. Kowalik, J. Quarles, B. Stevenson, and R. Taylor (ed.), Current protocols in microbiology. John Wiley & Sons, Hoboken, NJ. [DOI] [PMC free article] [PubMed]
- 35.Muffler, A., D. Fischer, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 101143-1151. [DOI] [PubMed] [Google Scholar]
- 36.Muffler, A., D. D. Traulsen, D. Fischer, R. Lange, and R. Hengge-Aronis. 1997. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 179297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 2821494-1497. [DOI] [PubMed] [Google Scholar]
- 38.Mulvey, M. A., J. D. Schilling, and S. J. Hultgren. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 694572-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulvey, M. A., J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bad bugs and beleaguered bladders: interplay between uropathogenic Escherichia coli and innate host defenses. Proc. Natl. Acad. Sci. USA 978829-8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson, C. N., V. J. Carabetta, T. Chowdhury, and T. J. Silhavy. 2006. LrhA regulates rpoS translation in response to the Rcs phosphorelay system in Escherichia coli. J. Bacteriol. 1883175-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rama, G., D. K. Chhina, R. S. Chhina, and S. Sharma. 2005. Urinary tract infections—microbial virulence determinants and reactive oxygen species. Comp. Immunol. Microbiol. Infect. Dis. 28339-349. [DOI] [PubMed] [Google Scholar]
- 43.Redford, P., P. L. Roesch, and R. A. Welch. 2003. DegS is necessary for virulence and is among extraintestinal Escherichia coli genes induced in murine peritonitis. Infect. Immun. 713088-3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redford, P., and R. A. Welch. 2006. Role of sigma E-regulated genes in Escherichia coli uropathogenesis. Infect. Immun. 744030-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robertson, G. T., and R. M. Roop, Jr. 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34690-700. [DOI] [PubMed] [Google Scholar]
- 46.Sittka, A., V. Pfeiffer, K. Tedin, and J. Vogel. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 63193-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 1831997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slonim, L. N., J. S. Pinkner, C. I. Branden, and S. J. Hultgren. 1992. Interactive surface in the PapD chaperone cleft is conserved in pilus chaperone superfamily and essential in subunit recognition and assembly. EMBO J. 114747-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonnleitner, E., S. Hagens, F. Rosenau, S. Wilhelm, A. Habel, K. E. Jager, and U. Blasi. 2003. Reduced virulence of a hfq mutant of Pseudomonas aeruginosa O1. Microb. Pathog. 35217-228. [DOI] [PubMed] [Google Scholar]
- 50.Soto, S. M., A. Smithson, J. P. Horcajada, J. A. Martinez, J. P. Mensa, and J. Vila. 2006. Implication of biofilm formation in the persistence of urinary tract infection caused by uropathogenic Escherichia coli. Clin. Microbiol. Infect. 121034-1036. [DOI] [PubMed] [Google Scholar]
- 51.Stanley, N. R., and B. A. Lazazzera. 2004. Environmental signals and regulatory pathways that influence biofilm formation. Mol. Microbiol. 52917-924. [DOI] [PubMed] [Google Scholar]
- 52.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7140-144. [DOI] [PubMed] [Google Scholar]
- 53.Svensson, L., B. I. Marklund, M. Poljakovic, and K. Persson. 2006. Uropathogenic Escherichia coli and tolerance to nitric oxide: the role of flavohemoglobin. J. Urol. 175749-753. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, K. M., V. A. Rhodius, and S. Gottesman. 2007. σE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 1894243-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Udekwu, K. I., F. Darfeuille, J. Vogel, J. Reimegard, E. Holmqvist, and E. G. Wagner. 2005. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 192355-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 511525-1533. [DOI] [PubMed] [Google Scholar]
- 57.Woolford, G., R. J. Casselden, and C. L. Walters. 1972. Gaseous products of the interaction of sodium nitrite with porcine skeletal muscle. Biochem. J. 13082P-83P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wright, K. J., P. C. Seed, and S. J. Hultgren. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 737657-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, A., S. Altuvia, A. Tiwari, L. Argaman, R. Hengge-Aronis, and G. Storz. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 176061-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 911-22. [DOI] [PubMed] [Google Scholar]
- 61.Zucht, H. D., J. Grabowsky, M. Schrader, C. Liepke, M. Jurgens, P. Schulz-Knappe, and W. G. Forssmann. 1998. Human beta-defensin-1: a urinary peptide present in variant molecular forms and its putative functional implication. Eur. J. Med. Res. 3315-323. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.