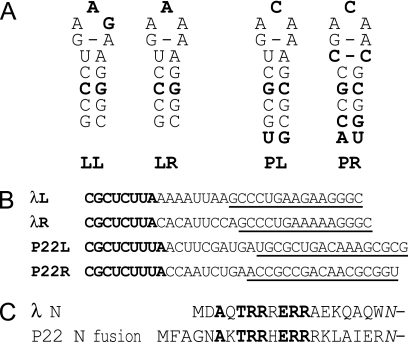
FIG. 1.
Comparison of λ and P22 nut site RNAs and N protein RNA-binding domains. (A) The secondary structures of boxBs found in bacteriophages λ and P22 are represented from the beginning of the boxB stem. LL, λ boxBleft; LR, λ boxBright; PL, P22 boxBleft; PR, P22 boxBright. Lightface nucleotides are found in either λ or P22 boxB sequences, while boldface nucleotides are found only in λ boxBs or only in P22 boxBs. The C:C pair adjacent to the loop of P22 boxBright is presumed to be stacked into the helix, perhaps as a hydrogen-bonded, nonstandard base pair. (B) Sequences of λ and P22 nut sites, where boxA is bold and boxB is underlined. λL, λ nutleft; λR, λ nutright; P22L, P22 nutleft; P22R, P22 nutright. In this study, all boxBs were placed in the context of λ nutleft to limit the effect of differences between boxAs and the boxA-boxB interval. (C) The sequences of the RNA-binding domains of λ N and the P22 N fusion plasmid used in this study are shown aligned from their amino termini to the fusion point with the common remainder of λ N. The italicized asparagine at the far right replaces a wild-type lysine in λ N to allow a restriction enzyme site in the N-supplying plasmids. Amino acids shown in bold are conserved between λ and P22 N.