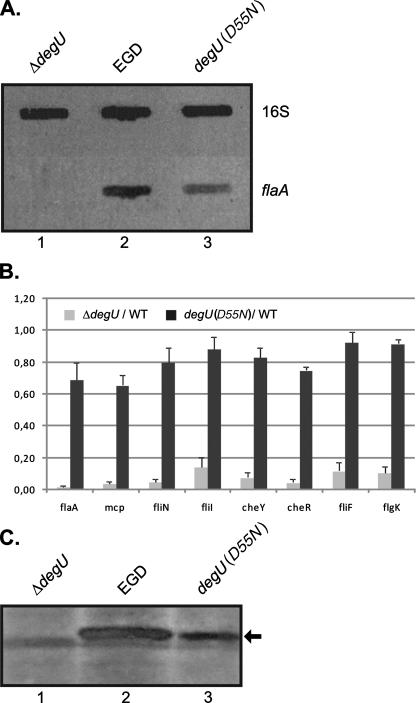
FIG. 2.
Expression analysis of motility genes in the L. monocytogenes EGD, ΔdegU, and degU(D55N) strains. (A) Slot blot hybridization carried out with equal amounts of RNA extracted from the L. monocytogenes EGD (lane 2), ΔdegU (lane 1), and degU(D55N) (lane 3) strains. Hybridization was performed with flaA- and 16S rRNA-specific probes on RNA from three independent preparations. The 16S rRNA-specific hybridization probe was PCR amplified with primer pair 16S-S5/16S-S3. The results of a representative experiment are shown. (B) The relative changes in transcription of flaA, fliN, fliI, fliF, flgK, cheY, cheR, and mcp in the L. monocytogenes ΔdegU and degU(D55N) mutant strains, compared to the wild-type strain EGD (WT), were assessed by qRT-PCR. The ratios of the transcript amount detected in the respective mutant to that of the wild type (ratio, mutant/WT) are depicted. The indicated ratios represent the means of results of three qRT-PCR experiments performed in duplicate with cDNA which was reverse transcribed from independent RNA preparations extracted from bacteria grown at 24°C. The relative transcription of the genes under study was normalized to that of the housekeeping gene rpoB, as described by Mertins et al. (5). The primer pairs generating the respective amplicons are listed in Table 1. Error bars indicate the standard deviations from the means. (C) Western blot analysis was performed with equal amounts of supernatant proteins prepared from liquid cultures of the L. monocytogenes EGD (lane 2), ΔdegU (lane 1), and degU(D55N) (lane 3) strains grown at 24°C using a polyclonal rabbit antiserum directed against flagella of L. monocytogenes serotype 1/2a. FlaA is indicated by an arrow. The results of one of three independent experiments are shown.