Abstract
Heparin-binding hemagglutinin (HBHA) is a virulence factor of tuberculosis which is responsible for extrapulmonary dissemination of this disease. A thorough biochemical characterization of HBHA has provided experimental evidence of a coiled-coil nature of HBHA. These data, together with the low-resolution structures of a full-length form and a truncated form of HBHA obtained by small-angle X-ray scattering, have unambiguously indicated that HBHA has a dimeric structure with an elongated shape.
Mycobacterium tuberculosis is one of the most devastating human microbial pathogens. It invades and multiplies in both phagocytes and epithelial cells (15). Adherence to host cells is mediated in bacteria by a plethora of fibrous systems, such as pili, fimbriae, and capsules (2, 23, 29). In M. tuberculosis, adherence to epithelial cells is mediated predominantly by the heparin-binding hemagglutinin adhesin (HBHA) (14, 18). This protein binds to heparan sulfate proteoglycans on the surface of epithelial cells and is therefore responsible for extrapulmonary dissemination of tuberculosis (19, 20). Binding to target epithelial cells involves the C-terminal lysine-rich domain of the protein, which is exposed at the mycobacterial cell surface (16). Like other bacterial adhesins, HBHA also expresses hemagglutination activity. This process has been proposed to be due to interactions between HBHA molecules on the surface of mycobacteria (4, 16). In particular, hemagglutination has been correlated with the ability of the N-terminal region of HBHA to form multimers (4, 28). However, despite the interest in HBHA both as a potential antigen against tuberculosis (13, 17) and as a diagnostic tool (6), no structural data are so far available, nor has detailed information on the protein oligomerization state yet been provided. We here present a biochemical characterization and the low-resolution structure determined by small-angle X-ray scattering (SAXS) of both the full-length HBHA and a truncated form, here denoted HBHAΔC, which lacks the C-terminal heparin-binding domain (residues 161 to 199). The results presented here provide clues to the molecular bases of bacterial agglutination processes.
Methods. (i) Cloning, expression, and purification of proteins.
The genes coding for HBHA and HBHAΔC (lacking residues 161 to 199) were amplified from the M. tuberculosis (strain H37Rv) genome by PCR and introduced into the pET-28a(+) (Novagen) and pETM-11 expression vectors, respectively. Sequenced clones were transformed in the Escherichia coli BL2(DE3) strain. Cells were grown to an optical density at 600 nm of approximately 0.5 in LB medium supplemented with kanamycin (50 μg/ml) at 37°C and were induced by 1 mM isopropyl-β-d-thiogalactoside (IPTG) for 5 h.
HBHA was purified on a heparin Hi-trap column (GE Healthcare) (10 mM Na phosphate, pH 7.0). The protein was eluted with a 0 to 1 M NaCl linear gradient. The proteic sample was further purified by size exclusion chromatography on Superdex 200 (GE Healthcare) (50 mM Tris-HCl,100 mM NaCl, pH 8.0). HBHAΔC protein was purified by Ni-nitrilotriacetic acid affinity chromatography (GE Healthcare) (equilibrated with 10 mM imidazole, 300 mM NaCl, and 50 mM Tris-HCl, pH 8.0) and eluted with a linear gradient of imidazole (10 to 300 mM). Following histidine tag removal, the protein was further purified by size exclusion chromatography on Superdex 200 (GE Healthcare).
(ii) Cross-linking experiments.
Cross-linking analysis of HBHAΔC was performed by incubating 1.2 mg ml−1 HBHAΔC in 20 mM HEPES buffer (pH 7.5), with either glutaraldehyde (0.3 to 2.3%) for 5 min at 37°C or bis-sulfosuccinnimidyl-suberate (BS3) (protein/BS3 ratios of 1:10 to 1:30) for 30 min at 25°C. Samples were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
(iii) CD experiments.
Circular dichroism (CD) spectra were recorded in 10 mM Na phosphate buffer (pH 7.0) in the wavelength range of 185 to 260 nm using a Jasco J-810 spectropolarimeter equipped with a Peltier thermostatic cell holder. Heat- and urea-induced denaturations were investigated by recording the CD signal at 222 nm. The final concentration of urea was in the range of 0.0 to 5.5 M, and each sample was incubated overnight at 293 K.
(iv) SAXS experiments.
Synchrotron SAXS data were collected on the X33 beamline (24) at EMBL, DESY, Hamburg. Scattering patterns of truncated and full-length HBHA were measured at several solute concentrations ranging from 2.0 to 7.7 mg/ml, and the range of momentum transfer 0.6 < s < 5.0 nm−1 [s = 4π sin(θ)/λ, where 2θ is the scattering angle and λ = 0.15 nm is the X-ray wavelength] was covered. Data were processed using standard procedures by PRIMUS (10). The molecular masses of the solutes were estimated based on the excluded (Porod) volume, Vp (22). For proteins, Porod (i.e., hydrated) volumes in nm3 are approximately two to three times larger than those expected given the molecular masses in kDa. Low-resolution models of HBHA were generated by two alternative ab initio approaches. The program GASBOR (27) represents a protein by an assembly of dummy residues and uses simulated annealing to build a locally “chain-compatible” dummy residue model inside a sphere with the diameter of the maximum particle size, Dmax. The multiphase bead modeling program MONSA (21, 26) represents the particle as a collection of densely packed beads inside the same spherical search volume. To describe the structure of HBHA, each bead can be assigned either to the solvent (index = 0) or to the HBHAΔC part or the C terminus (index = 1 and 2, respectively). Starting from random phase indices, simulated annealing was employed to search for a model which simultaneously fits the data for HBHAΔC and the full-length protein. The results of multiple reconstructions were averaged by the package DAMAVER (9) and SUPCOMB (11). The latter program aligns two arbitrary low- or high-resolution models by minimizing a dissimilarity measure, NSD, which is computed from the distances between two bead or atomic models as defined in reference 11. Generally, NSD values close to unity indicate that the two models are similar. The program DAMAVER selects the most typical model (i.e., that having the lowest average NSD with all the other models of the set of superimposed structures).
HBHA secondary structure.
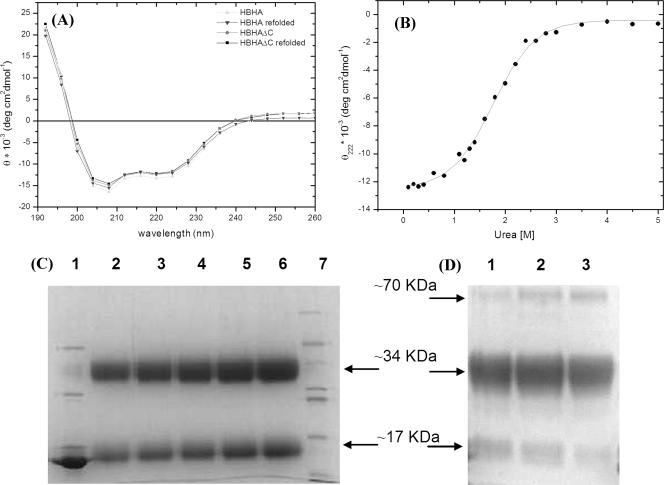
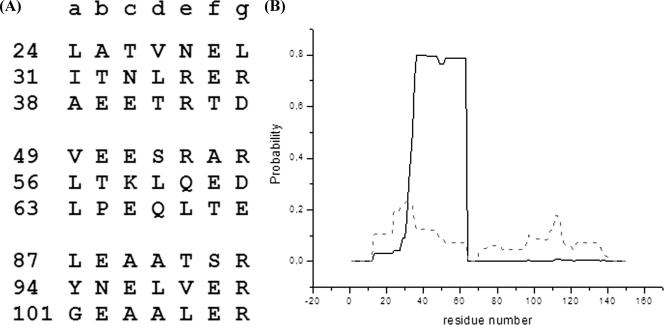
In order to experimentally assess the secondary structure content of HBHA, we performed CD experiments with both HBHA and HBHAΔC. The spectra of the two proteins are quite similar and are characterized by two negative broad peaks at about 208 and 220 nm (Fig. 1A). These spectra, with an α-helix band intensity at 208 nm stronger than that at 222 nm, are typical of proteins with a coiled-coil structure (1, 8, 12, 25). To validate this result, we performed chemical denaturation, using urea as a denaturating agent, of HBHAΔC. The urea denaturation profile of HBHAΔC is characteristic of a two-state helix-coil transition with a single infection point, Cm, centered at 1.8 M urea (Fig. 1B). This denaturation profile, with a peculiarly low urea Cm, has often been encountered in denaturation studies of coiled-coil systems (8). Thermal denaturation curves, recorded at 222 nm, show that denaturation occurs in one step (data not shown). Furthermore, folding of both proteins is fully reversible (Fig. 1A). This is consistent with the observation that in oligomeric structures, coiled coils often form in a single-step reaction, in which association and folding of peptide chains are tightly coupled (7). A coiled-coil propensity of HBHA is also predicted by sequence analysis (4, 27). Indeed, the HBHA sequence reveals the typical pattern of left-handed coiled coils, with a periodicity of seven (heptad repeats, abcdefg). As expected for left-handed coiled coils, residues in positions a and d are mainly hydrophobic, whereas those in positions e and g are mainly charged (Fig. 2). The highest propensity to form a coiled coil was calculated (using the program PCOILS [5]) for the region from residue 24 to 69, followed by the region from residue 87 to 107 (Fig. 2). The coiled-coil region from residue 24 to 69 was also predicted, using the program MULTICOIL (31), to have a high propensity to form two-stranded coiled coils (Fig. 2).
FIG. 1.
(A) CD curves of HBHA and HBHAΔC. CD curves after refolding are also shown. Melting temperatures are 37°C and 47°C for HBHA and HBHAΔC, respectively. (B) Urea-induced denaturation of HBHAΔC; experimental values are fitted with a sigmoidal curve. (C) SDS-PAGE analysis of HBHAΔC after cross-linking with glutaraldehyde. Lane 1 contains 50 μg of HBHAΔC as a control; lanes 2 to 6 contain 50 μg, 60 μg, 75 μg, 90 μg, and 100 μg of HBHAΔC, respectively; lane 7 contains molecular mass markers (66, 45, 36, 29, 24, 20, and 14.2 kDa). (D) SDS-PAGE analysis of HBHAΔC after cross-linking with BS3. Lanes 1 to 3 contain 30 μg HBHAΔC and protein/BS3 ratios of 1:10, 1:20, 1:30, respectively.
FIG. 2.
(A) Heptad repeats found in the HBHA sequence with the highest coiled-coil propensity, as predicted by the program PCOILS (5). Numbers on the left are amino acid residues. (B) Probability of residues forming either two-stranded (solid line) or three-stranded (dashed line) coiled coils, as predicted by the program MULTICOIL (31).
HBHA oligomerization state.
The evidenced coiled-coil nature of HBHA accords well with the hemagglutination function of HBHA, since coiled coils are folding motifs which guide oligomerization in a variety of systems (1, 12, 30). The oligomerization state of HBHA is not clear to date, as atomic force microscopy studies have indicated HBHA to be either dimeric or multimeric (28). Gel filtration profiles of freshly purified proteins showed the existence of species with molecular masses of 88 kDa for HBHA and 66 kDa for HBHAΔC. These estimates are compatible with a tetrameric arrangement of the proteins. However, since estimates of molecular masses by gel filtration depend on the protein shape, we decided to use two further approaches. First, we performed cross-linking experiments, followed by gel electrophoresis in denaturant conditions (15% SDS-PAGE), carried out using both glutaraldehyde and BS3 as cross-linking agents. These studies consistently indicated a molecular mass for HBHAΔC of 34 kDa, which corresponds to a dimer (Fig. 1C and D). Indeed, traces of mass with a molecular weight compatible with a tetrameric assembly (about 70 kDa) were detected only when a large excess of BS3 was used (Fig. 1D). The gel filtration profiles of cross-linked species were superposable with that of HBHAΔC, indicating that no unspecific cross-link had occurred. Cross-linking experiments were also performed with full-length HBHA and showed the existence of dimers, tetramers, and higher-order oligomers (not shown). The high-order oligomers were judged to be artifacts attributed to the lysine-rich C terminus (14 residues), which provides many cross-linking sites.
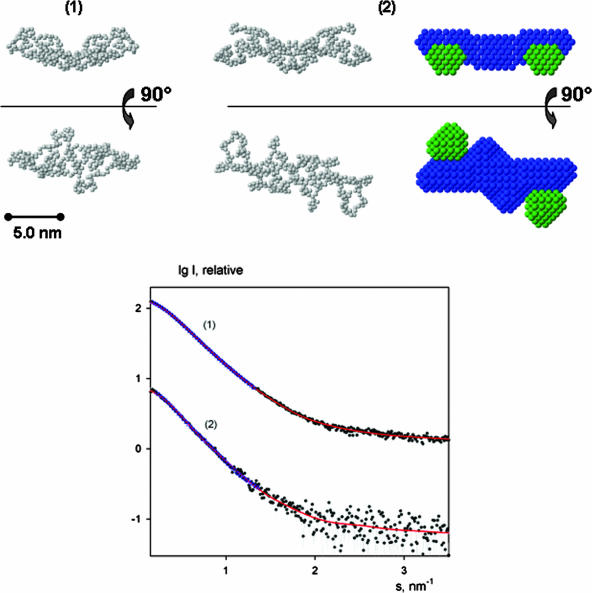
In order to further validate this result and to understand its inconsistency with results from gel filtration, we performed SAXS experiments with both HBHA and HBHAΔC. The scattering patterns from HBHA and HBHAΔC are presented in Fig. 3, and the structural parameters computed from the SAXS data are given in Table 1. The experimental Rg and Dmax values indicate that the particles are elongated, and the computed volumes are compatible with a dimeric organization of both constructs in solution. The increase of both the Rg and Dmax of the full-length protein compared to HBHAΔC suggests a peripheral location of the C terminus of HBHA.
FIG. 3.
SAXS modeling. (Top) Typical ab initio models of HBHAΔC (1) and full-length HBHA (2) generated using GASBOR (left and middle, respectively) and MONSA (right). In the MONSA representation, beads corresponding to the C termini are colored in green whereas the rest of the molecule is colored in blue. (Bottom) SAXS profiles from HBHAΔC (1) and the full-length protein (2). The experimental data are denoted by dots, and fits from dummy residue and bead models are given as solid red lines and blue triangles, respectively. The curves are in logarithmic scale for better visualization.
TABLE 1.
Overall SAXS parametersa
| Protein | Rg (nm)b | Dmax (nm) | Vp (nm3)b | χ | χmon | χdim | χtrim | χtetr |
|---|---|---|---|---|---|---|---|---|
| HBHAΔC | 3.36 ± 0.05 | 12.0 | 69 ± 10 | 1.24 | 1.32 | 1.11 | 1.47 | 2.25 |
| HBHA | 3.73 ± 0.12 | 13.5 | 107 ± 10 | 1.44 | 1.41 | 1.34 | 1.41 | 1.55 |
Rg, Dmax, and Vp are, respectively, the radius of gyration, maximum size, and excluded volume, calculated from the scattering data. χ is the discrepancy between the experimental data and computed scattering curves from the typical model reconstructed by MONSA. χmon, χdim, χtrim, and χtetr are typical discrepancies obtained in GASBOR reconstructions assuming the monomer, dimer, trimer, and tetramer organizations, respectively.
Mean ± standard deviation.
Elongated structure of HBHA dimer determined by SAXS.
As an additional check of the HBHA oligomeric state, several ab initio dummy residue reconstructions of both HBHA and HBHAΔC were performed by GASBOR (27), assuming monomeric, dimeric, trimeric, and tetrameric organizations. For dimers and tetramers the models were built assuming no symmetry and also with P2 and P222 (tetramers only) symmetry constraints. In all cases, the best agreement with the experimental data was achieved when dimers were assumed (Table 1). GASBOR modeling attempts with dimeric HBHA and HBHAΔC (both assuming P2 symmetry and without symmetry restrictions) yielded reproducible models (with an average NSD between the reconstructions of ∼1.5) whereby the restored shapes of HBHAΔC could be well accommodated within those for HBHA. This allowed us to assume the same conformation of the fragment from residue 1 to 160 in the truncated and full-length proteins and to fit the two data sets simultaneously by MONSA using twofold symmetry. A typical MONSA model (Fig. 3) agrees well with GASBOR dimeric models and neatly fits the experimental data (Table 1). All reconstructed models consistently show that HBHA displays an elongated shape with the C termini located at the outer ends (Fig. 3).
Conclusions.
We present here the first structural characterization of HBHA from M. tuberculosis. We have cloned, expressed, and purified both a full-length form and a truncated form of HBHA and applied various biophysical techniques to study its molecular properties and oligomerization, a key event for bacterial hemagglutination. CD studies have provided experimental evidence that HBHA presents a coiled-coil domain (Fig. 1). In addition, SAXS and cross-linking experiments have unambiguously shown that HBHA exists in solution as a dimer with an elongated shape (Fig. 3). SAXS results also indicate a peripheral location of the C-terminal positively charged arms of HBHA (Fig. 3). The protrusion of C termini toward the same side of the protein is consistent with the role attributed to the HBHA C terminus in driving the binding of bacteria by establishing electrostatic interactions with epithelial heparan sulfates (18). Overall, our data suggest that, like for other known systems (1, 3), the coiled-coil nature of the N-terminal region (residues 25 to 135) of HBHA is responsible for dimerization, which may be regarded as a process that leads to enhanced affinity to epithelial cells and improves cell adhesion.
Acknowledgments
This work was supported by the Italian Human ProteomeNet. M.V.P. and D.I.S. acknowledge support from the EU design study SAXIER (contract RIDS no. 011934).
We thank G. D'Auria for discussion.
Footnotes
Published ahead of print on 25 April 2008.
REFERENCES
- 1.Alfadhli, A., E. Steel, L. Finlay, H. P. Bachinger, and E. Barklis. 2002. Hantavirus nucleocapsid protein coiled-coil domains. J. Biol. Chem. 27727103-27108. [DOI] [PubMed] [Google Scholar]
- 2.Alteri, C. J., J. Xicohtencatl-Cortes, S. Hess, G. Caballero-Olin, J. A. Giron, and R. L. Friedman. 2007. Mycobacterium tuberculosis produces pili during human infection. Proc. Natl. Acad. Sci. USA 1045145-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, P., C. Schaffitzel, I. Berger, N. Ban, and U. Suter. 2003. Membrane association of myotubularin-related protein 2 is mediated by a pleckstrin homology-GRAM domain and a coiled-coil dimerization module. Proc. Natl. Acad. Sci. USA 10012177-12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delogu, G., and M. J. Brennan. 1999. Functional domains present in the mycobacterial hemagglutinin, HBHA. J. Bacteriol. 1817464-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber, M., J. Soding, and A. N. Lupas. 2006. Comparative analysis of coiled-coil prediction methods. J. Struct. Biol. 155140-145. [DOI] [PubMed] [Google Scholar]
- 6.Hougardy, J. M., K. Schepers, S. Place, A. Drowart, V. Lechevin, V. Verscheure, A. S. Debrie, T. M. Doherty, J. P. Van Vooren, C. Locht, and F. Mascart. 2007. Heparin-binding-hemagglutinin-induced IFN-gamma release as a diagnostic tool for latent tuberculosis. PLoS One 2e926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jelesarov, I., and H. R. Bosshard. 1996. Thermodynamic characterization of the coupled folding and association of heterodimeric coiled coils (leucine zippers). J. Mol. Biol. 263344-358. [DOI] [PubMed] [Google Scholar]
- 8.Kammerer, R. A., and M. O. Steinmetz. 2006. De novo design of a two-stranded coiled-coil switch peptide. J. Struct. Biol. 155146-153. [DOI] [PubMed] [Google Scholar]
- 9.Konarev, P. V., M. V. Petoukhov, V. V. Volkov, and D. I. Svergun. 2006. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 39277-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konarev, P. V., V. V. Volkov, A. V. Sokolova, M. H. J. Koch, and D. I. Svergun. 2003. PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36277-1282. [Google Scholar]
- 11.Kozin, M. B., and D. I. Svergun. 2001. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 3433-41. [Google Scholar]
- 12.Liu, J., Q. Zheng, Y. Deng, C. S. Cheng, N. R. Kallenbach, and M. Lu. 2006. A seven-helix coiled coil. Proc. Natl. Acad. Sci. USA 10315457-15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Locht, C., J. M. Hougardy, C. Rouanet, S. Place, and F. Mascart. 2006. Heparin-binding hemagglutinin, from an extrapulmonary dissemination factor to a powerful diagnostic and protective antigen against tuberculosis. Tuberculosis (Edinburgh) 86303-309. [DOI] [PubMed] [Google Scholar]
- 14.Menozzi, F. D., R. Bischoff, E. Fort, M. J. Brennan, and C. Locht. 1998. Molecular characterization of the mycobacterial heparin-binding hemagglutinin, a mycobacterial adhesin. Proc. Natl. Acad. Sci. USA 9512625-12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menozzi, F. D., V. M. Reddy, D. Cayet, D. Raze, A. S. Debrie, M. P. Dehouck, R. Cecchelli, and C. Locht. 2006. Mycobacterium tuberculosis heparin-binding haemagglutinin adhesin (HBHA) triggers receptor-mediated transcytosis without altering the integrity of tight junctions. Microbes Infect. 81-9. [DOI] [PubMed] [Google Scholar]
- 16.Menozzi, F. D., J. H. Rouse, M. Alavi, M. Laude-Sharp, J. Muller, R. Bischoff, M. J. Brennan, and C. Locht. 1996. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 184993-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parra, M., T. Pickett, G. Delogu, V. Dheenadhayalan, A. S. Debrie, C. Locht, and M. J. Brennan. 2004. The mycobacterial heparin-binding hemagglutinin is a protective antigen in the mouse aerosol challenge model of tuberculosis. Infect. Immun. 726799-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pethe, K., S. Alonso, F. Biet, G. Delogu, M. J. Brennan, C. Locht, and F. D. Menozzi. 2001. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature 412190-194. [DOI] [PubMed] [Google Scholar]
- 19.Pethe, K., M. Aumercier, E. Fort, C. Gatot, C. Locht, and F. D. Menozzi. 2000. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J. Biol. Chem. 27514273-14280. [DOI] [PubMed] [Google Scholar]
- 20.Pethe, K., P. Bifani, H. Drobecq, C. Sergheraert, A. S. Debrie, C. Locht, and F. D. Menozzi. 2002. Mycobacterial heparin-binding hemagglutinin and laminin-binding protein share antigenic methyllysines that confer resistance to proteolysis. Proc. Natl. Acad. Sci. USA 9910759-10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petoukhov, M. V., and D. I. Svergun. 2006. Joint use of small-angle X-ray and neutron scattering to study biological macromolecules in solution. Eur. Biophys. J. 35567-576. [DOI] [PubMed] [Google Scholar]
- 22.Porod, G. 1982. General theory, p. 17-51. In O. Kratky (ed.), Small-angle X-ray scattering. Academic Press, London, United Kingdom.
- 23.Remaut, H., R. J. Rose, T. J. Hannan, S. J. Hultgren, S. E. Radford, A. E. Ashcroft, and G. Waksman. 2006. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol. Cell 22831-842. [DOI] [PubMed] [Google Scholar]
- 24.Roessle, M. W., R. Klaering, U. Ristau, B. Robrahn, D. Jahn, T. Gehrmann, P. Konarev, A. Round, S. Fiedler, C. Hermes, and D. Svergun. 2007. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J. Appl. Crystallogr. 40s190-s194. [Google Scholar]
- 25.Steinmetz, M. O., I. Jelesarov, W. M. Matousek, S. Honnappa, W. Jahnke, J. H. Missimer, S. Frank, A. T. Alexandrescu, and R. A. Kammerer. 2007. Molecular basis of coiled-coil formation. Proc. Natl. Acad. Sci. USA 1047062-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svergun, D. I. 1999. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 762879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svergun, D. I., M. V. Petoukhov, and M. H. J. Koch. 2001. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 802946-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verbelen, C., V. Dupres, D. Raze, F. Dewitte, C. Locht, and Y. F. Dufrene. 2007. Single-molecule force spectroscopy of mycobacterial adhesin-adhesin interactions. J. Bacteriol. 1898801-8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vitagliano, L., A. Ruggiero, C. Pedone, and R. Berisio. 2007. A molecular dynamics study of pilus subunits: insights into pilus biogenesis. J. Mol. Biol. 367935-941. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289366-373. [DOI] [PubMed] [Google Scholar]
- 31.Wolf, E., P. S. Kim, and B. Berger. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 61179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]