Abstract
ResA is an extracytoplasmic membrane-bound thiol-disulfide oxidoreductase required for cytochrome c maturation in Bacillus subtilis. Previous biochemical and structural studies have revealed that the active-site cysteinyls cycle between oxidized and reduced states with a low reduction potential and that, upon reduction, a hydrophobic cavity forms close to the active site. Here we report in vivo studies of ResA-deficient B. subtilis complemented with a series of ResA variants. Using a range of methods to analyze the cellular cytochrome c content, we demonstrated (i) that the N-terminal transmembrane segment of ResA serves principally to anchor the protein to the cytoplasmic membrane but also plays a role in mediating the activity of the protein; (ii) that the active-site cysteines are important for cytochrome c maturation activity; (iii) that Pro141, which forms part of the hydrophobic cavity and which adopts a cis conformation, plays an important role in protein stability; (iv) that Glu80, which lies at the base of the hydrophobic cavity, is important for cytochrome c maturation activity; and, finally, (v) that Pro141 and Glu80 ResA mutant variants promote selective maturation of low levels of one c-type cytochrome, subunit II of the cytochrome c oxidase caa3, indicating that this apocytochrome is distinct from the other three endogenous c-type cytochromes of B. subtilis.
c-type cytochromes have diverse functions within cells. For example, they are components of respiratory and photosynthetic electron transfer chains (32), and one is an apoptosis-triggering factor in eukaryotes (44). These cytochromes are distinct from other cytochromes in that their heme group(s) is covalently attached to the protein via (usually) two thioether bonds between the heme vinyl groups and the side chains of two cysteine residues located within a conserved CXXCH sequence motif (29).
The structure and function of many c-type cytochromes have been characterized in detail. In contrast, relatively little is known about how these molecules acquire their covalently bound heme, a posttranslational process commonly referred to as cytochrome c maturation (CCM). Although it has been shown that this reaction can occur spontaneously under mild conditions in the presence of reduced apocytochrome and reduced heme (11), in all known natural cases a specific cellular machinery with variable complexity is required in vivo. To date, three markedly different CCM systems have been identified (2, 24, 32, 38). The most complex of these systems is system I, which comprises eight or nine specific proteins and which operates in mitochondria of plants and protozoans and in alpha- and gammaproteobacteria, such as Paracoccus denitrificans and Escherichia coli. System II is apparently simpler as it comprises only three specific proteins and is particularly widespread; it is present in plant chloroplasts, cyanobacteria, and gram-positive bacteria, including Bacillus subtilis (15, 26), as well as beta-, delta-, and epsilonproteobacteria, such as Bordetella pertussis and Helicobacter pylori (17). System III, which operates in mitochondria of many eukaryotes, is the simplest system, comprising a heme lyase (13) and possibly a reduction pathway (7). In addition to these well-established systems, there is now evidence for a fourth system in the Trypanosomatidae, which contain c-type cytochromes with only a single cysteine within the heme-binding motif (XXXCH) (3). Although these systems share common features, it is not clear why such different systems have evolved.
B. subtilis, the best-characterized gram-positive bacterium, contains four c-type cytochromes, all of which are membrane bound: CccA (cytochrome c550) (43), CccB (cytochrome c551) (5), QcrC (the cytochrome c subunit of the bc complex) (46), and CtaC (subunit II of the cytochrome c oxidase caa3) (42). Studies of the maturation of c-type cytochromes of B. subtilis, which are not required for growth or sporulation (37), have led to the identification of ResA, ResB, and ResC as essential components of CCM system II. ResB and ResC are integral membrane proteins that likely function in transmembrane heme transport and in catalyzing the covalent attachment reaction (26). ResA is an extracytoplasmic membrane-anchored thioredoxin-like protein which is believed to specifically reduce the CXXCH motif of apocytochromes c prior to heme attachment (15, 27). Reducing equivalents for the ResA-mediated reduction are supplied by another integral membrane protein, CcdA, which functions as a general transmembrane electron transfer protein required for sporulation, as well as CCM (35-37).
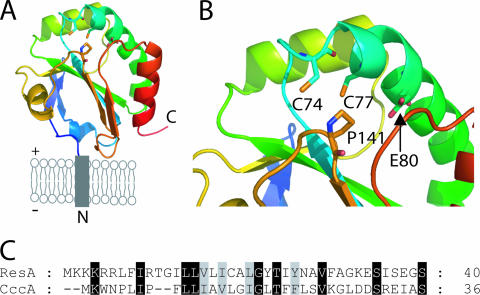
The high-resolution structure of oxidized ResA revealed a classical thioredoxin fold, comprising a mixed four-strand β-sheet surrounded by three α-helices, with two additional structural elements: a β-hairpin at the N terminus and a central insertion comprising a helix and loop region (Fig. 1). The structure of the reduced protein revealed a net loss of stabilizing interactions at the active site (consistent with the low potential determined for soluble ResA [9, 10]) and a number of concerted conformational rearrangements leading to the formation of a hydrophobic cavity with a polar base, which is proposed to be important for recognition of the presumed apocytochrome c substrates.
FIG. 1.
Structural properties of B. subtilis ResA. (A) Cartoon representation of ResA in the cytoplasmic membrane. The plus and minus signs indicate the outside and cytoplasmic side of the membrane, respectively, and N and C indicate the amino and carboxy termini of the protein, respectively. (B) Active-site and hydrophobic cavity regions of ResA. The images were generated using Pymol (12) and pdb file 1SU9. (C) Comparison of the amino acid residue sequences of the N-terminal anchors of ResA and CccA.
Here we describe an in vivo investigation of the functional importance of the specific sequence of the protein's membrane anchor, the active-site cysteinyls, and two residues, Glu80 and Pro141, located in the near-active-site hydrophobic cavity. The data indicate that the membrane anchor of ResA, although principally functioning to tether the soluble domain to the membrane, also has a role in mediating the transfer of electrons from ResA to its substrates. The data further demonstrate that the active-site cysteine residues are very important for CCM and, remarkably, that cells containing the mutant variants ResA(E80Q) and ResA(P141S/T) selectively mature only one c-type cytochrome, CtaC, indicating that this apocytochrome has different properties than the other three cytochromes. Pro141 is also shown to be important for the stability of ResA.
MATERIALS AND METHODS
Strains, growth media, and in vitro DNA techniques.
The bacterial strains and plasmids used are shown in Table 1. Escherichia coli strains were grown at 37°C in LB broth (33) or on LA plates consisting of LB with 1.5% (wt/vol) agar. B. subtilis strains were grown at 37°C in nutrient sporulation medium with phosphate (NSMP) (18), on plates containing NSMP with 1.5% agar, or on tryptose blood agar base plates. Antibiotics, where appropriate, were added to growth media at the following concentrations: for B. subtilis, 4 mg/liter chloramphenicol and 1 or 4 mg/liter erythromycin; and for E. coli, 100 mg/liter ampicillin. For B. subtilis strain LUL9 and its derivatives, isopropyl-β-d-thiogalactopyranoside (1 mM) was added to the growth medium (15).
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Genotype and/or relevant propertiesa | Origin and/or referenceb |
|---|---|---|
| B. subtilis strains | ||
| 1A1 | trpC2 | BGSCc |
| LUL9 | trpC2 resAΩpLLE36; Emr | 15 |
| LUN1 | trpC2 resAΩpLLE36 amyE::Pspac-resA; Emr Cmr | pALR12 → LUL9; this study |
| LUN2 | trpC2 resAΩpLLE36 amyE::Pspac-resA(C74A); Emr Cmr | pALR34 → LUL9; this study |
| LUN3 | trpC2 resAΩpLLE36 amyE::Pspac-resA(C77A); Emr Cmr | pALR35 → LUL9; this study |
| LUN4 | trpC2 resAΩpLLE36 amyE::Pspac-resA(C74A/C77A); Emr Cmr | pALR39 → LUL9; this study |
| LUN5 | trpC2 resAΩpLLE36 amyE::Pspac-resA(E80Q); Emr Cmr | pALR40 → LUL9; this study |
| LUN6 | trpC2 resAΩpLLE36 amyE::Pspac-resA(P141S); Emr Cmr | pCHN2 → LUL9; this study |
| LUN7 | trpC2 resAΩpLLE36 amyE::Pspac-resA(P141T); Emr Cmr | pCHN4 → LUL9; this study |
| E. coli strains | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′[traD36 proAB+ lacIqlacZΔM15] | 45 |
| MM294 | F−endA1 hsdR17(rk− mk−) supE44 thi relA1 mcrA Δ(mrr-hsdRMS-mcrBC) | 4 |
| Plasmids | ||
| pUC18 | Cloning vector; Apr | 45 |
| pUC19 | Cloning vector; Apr | 45 |
| pHP13 | Emr Cmr | 19 |
| pHP13Es | Cmr | 15 |
| pRAN1Es | pHP13Es containing 0.73-kb fragment with resA and its natural promoter; Cmr | 15 |
| pLUT191 | pUC19 with part of cccA on a 0.6-kb EcoRI-KpnI fragment; Apr | 36 |
| pLUDL01 | pUC19 with cccA′-resA′ encoding Cyt c″-ResA on a 1.1-kb EcoRI-BamHI fragment; Apr | This study |
| pLUJES1 | pHP13Es with cccA′-resA′ encoding Cyt c″-ResA on a 1.1-kb EcoRI-BamHI fragment; Cmr | This study |
| pVK48 | pDH32 derivative; PspaclacI; Apr Cmr | V. K. Charyd |
| pALR9 | pUC18 with wild-type resA blunt end cloned at SmaI site; Apr | This study |
| pALR12 | pVK48 with wild-type resA on a 0.59-kb XbaI/HindIII fragment under Pspac; Apr Cmr | This study |
| pALR33 | pUC18 with resA [encoding ResA(C74A)] at SmaI site; Apr | This study |
| pALR34 | pVK48 with resA [encoding ResA(C74A)] on a 0.59-kb XbaI/HindIII fragment under Pspac; Apr Cmr | This study |
| pALR32 | pUC18 with resA [encoding ResA(C77A)] at SmaI site; Apr | This study |
| pALR35 | pVK48 with resA [encoding ResA(C77A)] on a 0.59-kb XbaI/HindIII fragment under Pspac; Apr Cmr | This study |
| pALR36 | pUC18 with resA [encoding ResA(C74A/C77A)] at SmaI site; Apr | This study |
| pALR39 | pVK48 with resA [encoding ResA(C74A/C77A)]on a 0.59-kb XbaI/HindIII fragment under Pspac; Apr Cmr | This study |
| pALR37 | pUC18 with resA [encoding ResA(E80Q)] at SmaI site; Apr | This study |
| pALR40 | pVK48 with resA [encoding ResA(E80Q)] on a 0.59-kb XbaI/HindIII fragment under Pspac; Apr Cmr | This study |
| pCHN1 | pUC18 with resA [encoding ResA(P141S)] at SmaI site; Apr | This study |
| pCHN2 | pVK48 with resA [encoding ResA(P141S)] on a 0.59-kb XbaI/HindIII fragment under Pspac; Apr Cmr | This study |
| pCHN3 | pUC18 with resA [encoding ResA(P141T)] at SmaI site; Apr | This study |
| pCHN4 | pVK48 with resA [encoding ResA(P141T)] on a 0.59-kb XbaI/HindIII fragment under Pspac; Apr Cmr | This study |
Apr, resistance to ampicillin; Cmr, resistance to chloramphenicol; Emr, resistance to erythromycin.
An arrow indicates transformation of the strain.
BGSC, Bacillus Genetic Stock Center, Ohio.
V. K. Chary, Temple University School of Medicine.
General molecular genetics techniques described by Sambrook et al. (33) were used. Plasmid DNA was isolated using miniprep kits (Qiagen). Chromosomal DNA was isolated from B. subtilis strains as described by Marmur (28). E. coli was transformed using chemically competent cells (33), and B. subtilis was grown to competence as previously described (21).
Construction of a cccA′-resA′ fusion.
resA (nucleotides 90 to 537, encoding residues 30 to 179 of the wild-type protein) was PCR amplified using primers resA-5′ and resA-3′ and pRAN1Es (15) as the template. Table S1 in the supplemental material shows all of the primers used in this study. The PCR product was cut using BamHI/KpnI and ligated into pLUT191 cut with the same enzymes, generating pLUDL01, a pUC19-derived construct containing the promoter, ribosome binding sequence, and proximal end of the cccA gene (bp 18 to 683) fused to the distal part of resA (bp 90 to 537). pLUDL01 was digested with EcoRI/BamHI, and the cccA′-resA′ fragment was ligated into pHP13Es cut with the same enzymes, generating pLUJES1.
Cloning and site-directed mutagenesis of full-length ResA.
The wild-type full-length resA gene with its ribosome binding sequence was amplified from B. subtilis strain 1A1 chromosomal DNA using primers resA17 and resA18, which introduced a HindIII site at the start of the fragment and an XbaI site at the end. The PCR product was cloned into pUC18 digested with SmaI, generating pALR9. Site-directed mutagenesis was carried out using a whole-plasmid method (22) with pALR9 as the template to generate mutant resA genes encoding C74A (pALR33), C77A (pALR32), C74A/C77A (pALR36), E80Q (pALR37), P141S (pCHN1), and P141T (pCHN3) ResA variants. The XbaI/HindIII fragments of pALR9 and each of the mutant plasmids were cloned into pVK48 cut with the same enzymes, generating plasmids encoding wild-type ResA (pALR12) and C74A (pALR34), C77A (pALR35), C74A/C77A (pALR39), E80Q (pALR40), P141S (pCHN2), and P141T (pCHN4) variants. Each pVK48-derived plasmid was used to transform resA-defective B. subtilis strain LUL9 to chloramphenicol resistance. Transformants were tested for amylase activity on tryptose blood agar base plates containing 1% (wt/vol) starch using Lugol's solution, and the integration of resA at amyE was confirmed for each mutant strain by PCR analysis and by sequencing over the integration site (MWG Biotech). The resulting new strains are listed in Table 1.
Assays for cytochromes c.
B. subtilis strains deficient in cytochrome c oxidase caa3 were identified by testing for N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) oxidation, as previously described (26). The activity of cytochrome c oxidase caa3 was measured more quantitatively using a cytochrome c oxidase activity assay (42). Briefly, 6.9 mg of horse heart cytochrome c was dissolved in 20 mM Tris (pH 8) and reduced with 1 mM sodium dithionite. Excess reductant was removed, and the protein was exchanged into 20 mM 3-morpholinopropanesulfonate (MOPS) (pH 7.4) using a desalting column (PD10; GE Healthcare). Each membrane preparation was rapidly mixed with an equal volume of a reduced cytochrome c solution to obtain final concentrations of ∼40 μg of membrane protein per ml and 40 μM cytochrome c in 20 mM MOPS (pH 7.4), and the mixture was stirred continuously using a microstirrer bar. Changes in absorbance at 540 and 550 nm were measured every 15 s using a Perkin Elmer Lambda 800 spectrophotometer. Differences in absorbance (A550 − A540) were plotted against time, and the initial rate of cytochrome c oxidation was determined from data beginning at 30 s after mixing.
The total cytochrome c content of membranes was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and heme staining (41). Membrane samples (containing approximately 100 μg of protein) were run on an SDS-PAGE gel. The gel was soaked in 10% trichloroacetic acid for 10 min, washed twice (10 min each) in water, and stained for heme by addition of a 20-ml solution containing 20 mg o-dianisidine-HCl and 0.7% (vol/vol) H2O2 in 0.1 M sodium citrate (pH 4.4), followed by incubation at room temperature for 30 min. The reaction was stopped by washing the gel several times in water.
Other methods.
Western blot analysis was performed by transferring proteins separated by SDS-PAGE to polyvinylidene difluoride membranes using an electroblotter (Hoefer). Rabbit antiserum against ResA (15) was used as the primary antibody (at a 1,000-fold dilution), and horseradish peroxidase-conjugated donkey anti-rabbit serum was used as the secondary antibody (at a 5,000-fold dilution). Immunoreactive proteins were visualized using a peroxidase-reactive chemiluminescence detection system (GE Healthcare). SDS-PAGE was carried out using either the Laemmli (25) or Schägger-von Jagow (34) systems. Membranes were isolated from B. subtilis strains essentially as previously described (20) and were stored at −80°C until they were required. The protein concentrations of membrane fractions were determined using the bicinchoninic acid assay, with bovine serum albumin as the standard. For membrane preparations from cultures grown in the presence of 1 mM dithiothreitol (DTT), the reducing capacity of DTT was verified at the time of harvest using dinitro-5,5′-dithiodibenzoic acid (Ellman's reagent). Amino acid sequence alignment was performed using ClustalW (40), and alignments were edited using Genedoc (30). Digital images of gels were converted to grayscale and corrected for contrast using Adobe Photoshop Lite.
RESULTS
The membrane anchor of ResA plays a role in CCM.
ResA is located on the outside of the cytoplasmic membrane and is tethered to it by an N-terminal transmembrane segment, such that the N terminus of the protein is located in the cytoplasm. A soluble form of ResA has been structurally characterized (in both oxidized and reduced states), and the redox and pKa properties of the protein have been determined (8-10, 27). While it is unlikely that the membrane anchor significantly affects the physicochemical properties of the soluble domain, it is not clear whether the membrane anchor serves only to localize ResA to the correct cellular compartment. To investigate this, the membrane anchor segment of ResA was substituted for that of CccA, a small cytochrome c also anchored to the B. subtilis cytoplasmic membrane via a single transmembrane segment (43). A plasmid encoding the CccA-ResA fusion protein (pLUJES1) was introduced into the B. subtilis 1A1 (parental) and LUL9 (resA-deficient) strains. Plate colony TMPD staining of LUL9/pLUJES1 revealed that the fusion protein was active in CCM and that the intensity of staining was similar to that of the 1A1 strain (not shown). As expected, colonies of LUL9 containing an empty plasmid vector (pHP13Es) did not show TMPD oxidation activity (not shown). A more quantitative measure of cytochrome c oxidase activity was obtained by measuring the oxidation of cytochrome c directly using an in vitro assay with isolated membranes (Table 2). The data indicated that the fusion protein is less active than normal ResA, with oxidase activity that is ∼25% of the activity of the wild-type control (1A1) and ∼40% of the activity of LUL9 producing wild-type ResA from an equivalent pHP13Es-derived plasmid construct (pRAN1Es) (15). It is not clear why membranes from LUL9/pRAN1Es exhibit slightly lower activity than 1A1 wild-type membranes (Table 2). Whatever the exact cause of this, the difference is relatively small and was not apparent from our previous analysis of the entire cytochrome c contents of 1A1 and LUL9/pRAN1Es (15).
TABLE 2.
Cytochrome c oxidation activities of membranes from different B. subtilis strains
| Strain | ResA variant | Oxidation activity (nM min−1 mg protein−1)a |
|---|---|---|
| 1A1 | Wild type | 83 ± 4 |
| LUL9 | 5 ± 4 | |
| LUL9/pLUJES1 | CccA′-ResA′ fusion | 20 ± 2 |
| LUL9/pRAN1Es | Wild type | 50 ± 6 |
| LUN1 | Wild type | 82 ± 6 |
| LUN2 | C74A | 13 ± 1 |
| LUN3 | C77A | 4 ± 2 |
| LUN4 | C74A/C77A | 4 ± 1 |
| LUN5 | E80Q | 12 ± 3 |
| LUN6 | P141S | 15 ± 2 |
| LUN7 | P141T | 28 ± 4 |
The data are the means ± standard errors of several measurements (n ≥ 3) obtained using at least two different membrane preparations.
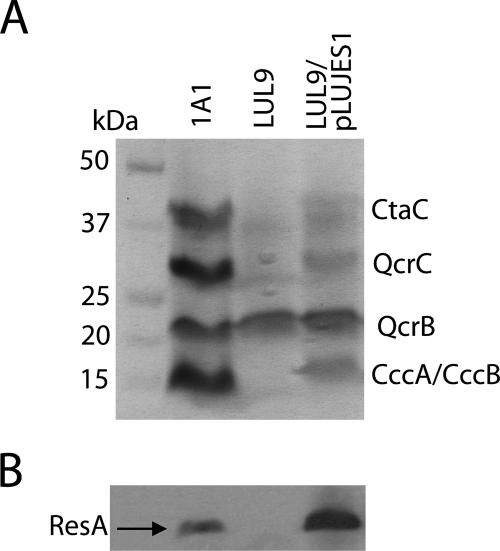
TMPD staining and the cytochrome c oxidase activity assay provide information only about the activity of the cytochrome c of cytochrome c oxidase, CtaC. To determine the levels of the other three cytochromes c (QcrC, CccA, and CccB) found in B. subtilis, membrane samples were analyzed by SDS-PAGE and heme staining (Fig. 2A). Bands corresponding to each of the c-type cytochromes of B. subtilis were detected in the wild-type control (1A1), and none of the system II-dependent c-type cytochromes were detected in LUL9. The band at ∼22 kDa was due to an unusual heme that is covalently attached to the cytochrome b subunit (QcrB) of the bc complex (46). Although the absolute level of heme staining was somewhat variable, membranes of LUL9/pLUJES1 exhibited levels of mature cytochromes that were significantly lower than those observed for the same concentration of membrane protein from a 1A1 preparation. Thus, the CCM deficiency was observed for all c-type cytochromes.
FIG. 2.
Cytochromes c and CccA-ResA fusion protein in membranes of different strains. (A) SDS-PAGE and heme staining of membranes from B. subtilis 1A1 (parental strain), LUL9 (lacking ResA), and LUL9/pLUJES1 (containing a plasmid-borne cccA′-resA′ fusion gene). Bands due to the c-type cytochromes and QcrB protein of B. subtilis are indicated on the right. (B) Western blot of membranes from the strains shown in panel A, probed with anti-ResA antiserum.
Because the fusion protein is produced from a multicopy plasmid, it was important to probe the overall levels of ResA protein present in the membrane preparations. A Western blot analysis of the membrane preparations using antiserum against the catalytic domain of ResA (15) was performed (Fig. 2B), which demonstrated that the CccA-ResA fusion protein was present at levels that were significantly higher than those in 1A1 preparations (as was also observed when ResA was overproduced in LUL9 from pRAN1Es [15]).
The active-site cysteinyls of ResA are important for CCM.
To test the importance of the active-site cysteine residues of ResA, wild-type full-length resA and mutants of this gene encoding single-cysteine variants (C74A and C77A) and double-cysteine variants (C74A/C77A) under the Pspac promoter were generated and integrated into the amyE gene of B. subtilis LUL9, which lacks a functional resA gene (Table 1).
TMPD staining of colonies of the resulting transformants revealed that the cytochrome caa3 activities of all of the strains with cysteine variants of ResA were very low. From this we concluded that the assembly of CtaC is significantly affected. To obtain a more quantitative indication of the extent of cytochrome caa3 activity (relative to the wild-type strain and LUL9 complemented with wild-type resA [LUN1]), the in vitro cytochrome c oxidase assay was employed (Table 2). LUL9 exhibited ∼10-fold-lower activity than 1A1 or LUN1. B. subtilis LUN2, which contained ResA(C74A), showed marginally higher activity than LUL9, while LUN3 and LUN4 with ResA(C77A) and ResA(C74A/C77A), respectively, exhibited activities similar to that of LUL9. Thus, the active-site cysteine residues of ResA are clearly important for the maturation of at least CtaC.
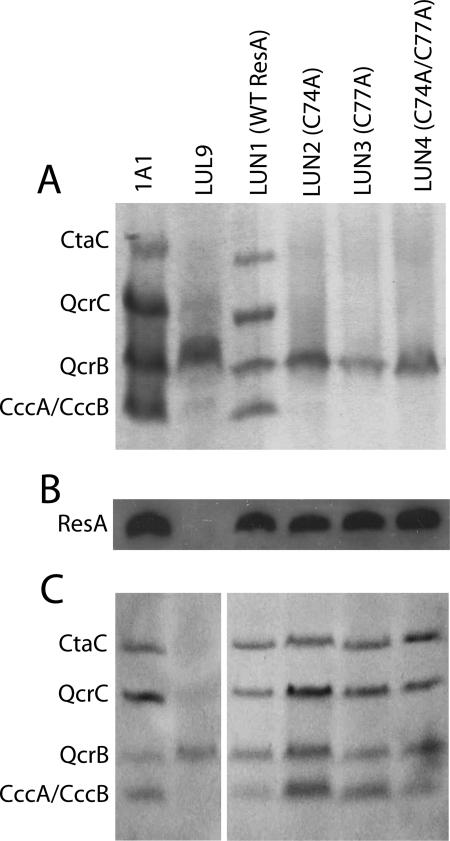
To determine the levels of the other three cytochromes c (QcrC, CccA, and CccB) found in B. subtilis, membrane samples were analyzed by SDS-PAGE and heme staining (Fig. 3). As in the experiment whose results are shown in Fig. 2, bands corresponding to each of the c-type cytochromes of B. subtilis were detected in the wild-type control and in LUN1, and none of the system II-dependent c-type cytochromes were detected in LUL9. Membranes from strains LUN2, LUN3, and LUN4 did not contain detectable levels of any of the c-type cytochromes, indicating that the ResA variants in which cysteinyls were replaced were inactive.
FIG. 3.
Cytochromes c and Cys mutant variants of ResA in membranes of different strains. (A) SDS-PAGE and heme staining of membranes from B. subtilis 1A1, LUL9, LUN1, LUN2, LUN3, and LUN4. Bands due to the c-type cytochromes of B. subtilis are indicated on the left. (B) Western blot of membranes from the strains shown in panel A, probed with anti-ResA antiserum. (C) Like panel A, except that membranes were purified from strains grown in the presence of 1 mM DTT.
To verify the presence of mutant ResA proteins in membranes of the strains, Western blot analyses were performed using anti-ResA antibodies (Fig. 3B). The results confirmed that the protein variants are indeed stable, consistent with the crystal structures of the soluble ResA(C74A), ResA(C77A), and ResA(C74A/C77A) proteins, which showed that, apart from the substituted residue, there are essentially no structural changes compared to the wild-type protein (27).
Previously, we showed that cytochrome c oxidase activity was restored when LUL9 was grown on NSMP agar containing 15 mM DTT (15). To determine whether all CCM was restored by the addition of DTT to the growth medium, membranes were prepared from cultures grown in the presence of 1 mM DTT. An SDS-PAGE and heme staining analysis of membrane preparations (Fig. 3C) revealed that CCM activity was restored in resA mutant strains LUN2 to LUN4. Activity was also restored in LUL9, although the intensities of bands corresponding to c-type cytochromes were lower than the intensities of the bands for 1A1 and LUN2 to LUN4. The data demonstrate that the active-site cysteines are not essential if an alternative source of electrons is provided. DTT can clearly access the outer side of the cytoplasmic membrane to fulfill this role.
Importance of ResA Pro141 and Glu80 for CCM.
Pro141 is absolutely conserved among thioredoxin-like thiol-disulfide oxidoreductases (TDORs). All available high-resolution structural data show that it occurs in a cis conformation, indicating its importance for the thiol-disulfide exchange reactions of TDORs. For example, in E. coli DsbA, Pro151 (equivalent to Pro141 in ResA) was shown to be important for interactions with both substrates and DsbB (which functions to reoxidize DsbA back to its active form); substitution with threonine led to the isolation of DsbA in complex (through a heterodisulfide) with numerous substrate proteins, and substitution with serine led to a stable DsbA-DsbB mixed disulfide complex (23). In order to investigate the importance of ResA Pro141 for CCM, resA mutants encoding ResA(P141T) and ResA(P141S) variants were generated and introduced into strain LUL9, as described above.
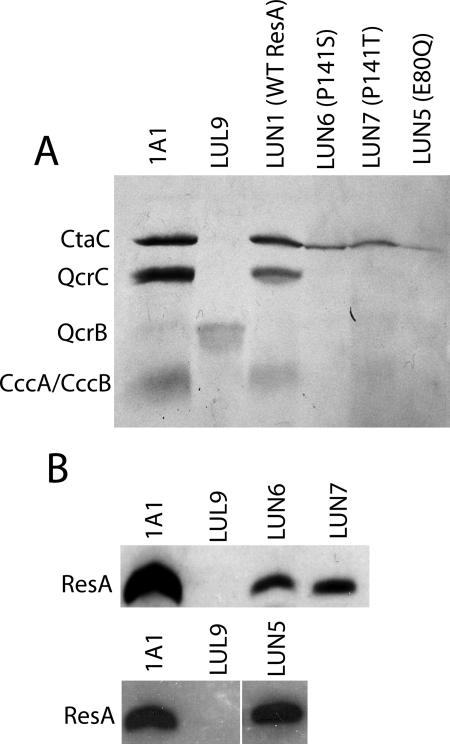
TMPD staining of colonies revealed levels of CtaC activity lower than the wild-type levels (not shown). Membranes isolated from LUN6 (P141S) and LUN7 (P141T) showed significantly reduced cytochrome c oxidase activities; the P141S variant resulted in <20% of the wild-type activity, and the P141T variant resulted in ∼35% of the wild-type activity (Table 2).
An SDS-PAGE and heme staining analysis of LUN6 and LUN7 membranes demonstrated that the ResA variants present in the two strains support maturation of CtaC, but at levels lower than those observed for 1A1 (Fig. 4A). Surprisingly, other c-type cytochromes are not matured to significant levels in LUN6 and LUN7. Membranes containing ResA(P141T) may contain low levels of CccA/CccB, but the levels are much lower than those in 1A1 membranes. Attempts to purify soluble ResA(P141S) produced in E. coli were not successful (A. Lewin and N. E. Le Brun, unpublished data), indicating that at least under the conditions tested, the variant ResA is not stable. Hence, it was important to establish whether the reduced activities detected were a consequence of the lack of the protein in isolated membranes or a consequence of a lack of activity due to the protein. Western blot analysis (Fig. 4B) showed that both proteins were present in membrane preparations, but at a low level (≤20% compared to the 1A1 control for wild-type ResA). No other immunoreactive bands were observed, indicating that under these conditions Pro141 variant ResA-substrate complexes are not stabilized. Furthermore, the level of ResA antigen in membrane preparations was found to decrease following a freeze-thaw cycle, consistent with the conclusion that the P141S and P141T ResA variants are not as stable as the wild-type protein or the cysteine variants described above.
FIG. 4.
Cytochromes c and Pro141 and Glu80 mutant variants of ResA in membranes of different strains. (A) SDS-PAGE and heme staining of membranes from B. subtilis 1A1, LUL9, LUN1, LUN5, LUN6, and LUN7. Bands due to the c-type cytochromes of B. subtilis are indicated on the left. We noted that the band due to QcrB (the cytochrome b subunit of the bc complex) is missing from each lane; the reason for this is not clear, but we have routinely observed variable staining of this protein, which contains an unusual covalently attached heme (46). (B) Western blot of membranes from the strains shown in panel A, probed with anti-ResA antiserum.
The structure of oxidized and reduced ResA revealed redox-dependent structural changes close to the active site that, upon reduction, result in the formation of a hydrophobic cavity near the active site with Glu80 at its base. This led to the proposal that the cavity, and Glu80 in particular, may play a role in substrate recognition (9, 10). This residue was recently shown to be very important in control of the acid-base properties of the ResA active-site cysteines. Replacement of Glu80 in soluble ResA by glutamine resulted in a significant decrease (>1 pH unit) in the pKa values of the cysteine side chains and in greater reactivity of the cysteine side chains with various reagents (27). Therefore, it was of interest to establish the in vivo activity of ResA bearing this substitution. A mutant resA gene was prepared as described above and integrated into the amyE locus of B. subtilis LUL9, generating strain LUN5. Again, TMPD staining of colonies indicated reduced cytochrome c activity (not shown), and the oxidase activity of isolated membranes was only ∼15% of the activity of 1A1 membranes (Table 2).
Analysis of the c-type cytochrome content of LUN5 membranes by SDS-PAGE and heme staining revealed that, like the Pro141 variants described above, ResA(E80Q) supports the maturation of only CtaC, albeit at a level that is less than the level observed for LUN1 (Fig. 4A). In contrast to the Pro141 variants, however, the ResA(E80Q) variant was detected at wild-type levels by Western blotting (Fig. 4B).
DISCUSSION
Replacement of the N-terminal transmembrane anchor of ResA by the equivalent membrane anchor of CccA resulted in a decrease in CCM activity. That the fusion protein contains a similar N-terminal sequence (Fig. 1C), which normally directs CccA to the cytoplasmic membrane, indicates that the fusion protein is correctly localized to the outside of the cytoplasmic membrane, and this conclusion is supported by the observation that there is CCM activity. However, the activity appears to be significantly lower than that of wild-type ResA and is also ∼60% lower than that observed for LUL9 complemented with plasmid-borne wild-type resA (pRAN1Es) expressed from the same promoter that was used to drive expression of the fusion protein, which provides a more precise control.
The membrane anchor in the fusion protein originates from a c-type cytochrome, and a possible complication in the interpretation of the data is that the anchor of CccA may be involved in the interaction of apo-CccA with the Res proteins during the heme attachment reaction. Thus, the fusion protein could block the interaction of apocytochromes, leading to reduced levels of CCM. However, it is very unlikely that the transmembrane anchors of c-type apocytochromes are important for interactions with the Res proteins because they have highly variable sequences, and the N termini of CccB and CtaC are processed to generate lipoproteins. In the case of CtaC, it is known that lipid modification and cleavage are not required for heme attachment (6). The only region with significant sequence similarity in all of the c-type cytochromes of B. subtilis occurs around the heme-binding motif (Fig. 5), and it is likely that this region determines the specificity of the interaction with the Res proteins. Thus, we concluded that the specific sequence of the membrane anchor of ResA has an influence on CCM activity. We noted that a similar conclusion about the role of the membrane anchor was drawn from studies of E. coli CcmG, the system I homologue of ResA, in which the natural membrane anchor was substituted for that of CcmE (1). The resulting fusion protein was produced at levels somewhat lower than the levels of the wild-type protein and resulted in approximately 50% CCM activity.
FIG. 5.
Comparison of the amino acid sequences of the heme domain of the four c-type cytochromes of B. subtilis. The complete sequences of CccA and CccB are aligned with the C-terminal parts of QcrC (residues 125 to 255) and CtaC (residues 151 to 356).
ResA reduces oxidized apocytochromes c prior to the covalent attachment of heme, a reaction catalyzed by ResB and/or ResC. It is possible, and perhaps even likely, that ResA interacts specifically with ResB and/or ResC to ensure that, once reduced, apocytochromes undergo the heme attachment reaction (rather than becoming reoxidized). Therefore, one possible explanation for the data reported here is that the membrane anchor plays a role in the recognition of ResB/ResC. Further investigation of the possible formation of a ResABC complex is necessary.
In E. coli, deletion of ccmG also resulted in a complete loss of CCM (16). Complementation of the ΔccmG strain with genes encoding active-site cysteine variants resulted in low but detectable levels of Bradyrhizobium japonicum cytochrome c550 (heterologously expressed from a separate plasmid within the cell), and these levels were increased by addition of l-cysteine to the growth medium (16). This led to the suggestion that in addition to TDOR activity, CcmG may have a role in stabilizing other proteins involved in CCM. Replacement of the active-site cysteine residues of ResA resulted in a loss of CCM activity in LUN3 [ResA(C77A)] and LUN4 [ResA(C74A/C77A)] and close to no activity in LUN2 [ResA(C74A)]. Thus, we concluded that the principal function of ResA lies in mediating thiol-disulfide exchange; ResA proteins which lack the ability to catalyze this process are unable to support significant CCM. This is consistent with the observation that the absence of ResA can be complemented by inactivation of BdbD, the DsbA homologue of B. subtilis, which is required for the formation of disulfide bonds on the outside of the cytoplasmic membrane (15). This showed that ResA is not required for CCM unless the apocytochrome substrates are in an oxidized state and that therefore ResA functions solely to reduce oxidized apocytochromes c prior to heme attachment. This conclusion is further supported by the observation that there is CCM activity in strains LUL9 and LUN2 to LUN4 when they are grown in the presence of DTT. The fact that the extent of maturation in membranes of LUL9 appeared to be lower than the extent of maturation for the other strains suggests that the ResA protein, which is absent in LUL9, may itself assist in the CCM process. The proposal that ResA interacts with ResB and ResC is consistent with this observation.
Pro141 lies close to the active site of ResA, forming part of the nearby hydrophobic cavity (Fig. 1). Replacement of Pro141 by serine or threonine was found to destabilize the protein fold; significant amounts of either Pro141 variant could be detected only in freshly prepared B. subtilis membranes, and the levels were depleted following a freeze-thaw cycle. These observations are consistent with the instability of a soluble version of the P141S variant and suggest that Pro141 is important for the stability of the protein fold. This contrasts with a report for E. coli CcmG, in which replacement of the cis-proline, P144, by an alanine resulted in a stable protein with an overall structure very similar to that of the wild-type protein (but with altered redox potential properties) (31).
The presence of small amounts of the Pro141 variants correlated well with low CtaC activity. Therefore, for CtaC, the normalized activity is similar to that of the wild-type protein, indicating that Pro141 does not appear to be important for the maturation of CtaC. Remarkably, none of the other c-type cytochromes were matured to detectable levels; that is, ResA(P141S/T) variants exhibit selective maturation of c-type cytochromes. Therefore, Pro141, in addition to its role in stabilizing the protein fold, also has a key role in the recognition of the QcrC, CccA, and CccB apocytochrome c substrates, as previously suggested by high-resolution structural studies (27).
Selective maturation of CtaC was also observed in membranes containing ResA(E80Q). Structural data previously indicated that the E80 residue, which lies at the base of the hydrophobic cavity juxtaposed to the active-site cysteine motif, is likely to be important in apocytochrome c recognition (9). Although the E80Q protein is like the wild type in terms of abundance and stability, only small amounts of mature CtaC were observed. Thus, ResA(E80Q) is deficient in the maturation of all c-type cytochromes. It exhibits some activity toward CtaC but essentially none toward QcrC, CccA, and CccB.
While it is clear that ResA is required for the reduction of apocytochromes c prior to covalent heme attachment and a model for substrate activation of ResA reductase activity has been proposed (27), a direct interaction with apocytochromes has not been conclusively demonstrated so far. The data reported here provide further evidence that ResA interacts directly with apocytochromes. If ResA served only to reduce an intermediate protein which subsequently reduced apocytochromes, we would not expect to observe selective maturation of cytochromes with different ResA variants.
Previous studies of the acid-base properties of a soluble form of ResA(E80Q) showed that the negative charge associated with Glu80 increases the pKa values of the active-site cysteinyl thiols, thus rendering the protein unreactive at physiological pH. This led to the proposal that partial masking of the negative charge of Glu80 may be important for activation of ResA. The low in vivo activity of the E80Q variant is consistent with an important role for this residue in substrate recognition/binding. Glu80 is highly conserved in thioredoxinlike extracytoplasmic TDORs, consistent with the hypothesis that it performs an important function in this class of protein. However, previous studies of E. coli CcmG in which the equivalent glutamate residue (Glu86) was replaced by alanine revealed levels of mature cytochrome c that were comparable to those in the wild-type strain (14). However, CcmG is not the terminal reductase in system I CCM; i.e., it occurs earlier in the transfer chain, accepting electrons from the transmembrane DsbD protein and passing them on to CcmH (39). Furthermore, the majority of extracytoplasmic TDORs in B. subtilis and E. coli do not function in CCM. Therefore, the role of Glu80 identified in ResA may well be peculiar to TDORs involved in the reduction of apocytochromes.
The data presented here indicate that Pro141 in ResA is critical for the stability of the protein and for stabilizing the interaction of ResA with the apocytochromes QcrC, CccA, and CccB, but not CtaC. Glu80 is not important for the stability of the protein, but it is important for the interaction with CtaC and essential for the interaction with the other apocytochromes. From the alignment of the four apocytochromes (Fig. 5) it is clear that CtaC is very different from the other apocytochromes in that it contains a series of inserts. In particular, it contains an insert immediately after its CXXCH heme attachment motif. These differences may be important for the selective maturation of CtaC observed here.
Supplementary Material
Acknowledgments
This work was supported by the United Kingdom EPSRC through award of a Ph.D. studentship to C.H., by BBSRC grant BB/C503597/1, and by Swedish Research Council grant 621-2007-6094.
We thank Dan Li for assistance with the construction of pLUJES1 and V. K. Chary for providing pVK48.
Footnotes
Published ahead of print on 2 May 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ahuja, U., and L. Thony-Meyer. 2006. The membrane anchors of the heme chaperone CcmE and the periplasmic thioredoxin CcmG are functionally important. FEBS Lett. 580216-222. [DOI] [PubMed] [Google Scholar]
- 2.Allen, J. W., O. Daltrop, J. M. Stevens, and S. J. Ferguson. 2003. C-type cytochromes: diverse structures and biogenesis systems pose evolutionary problems. Philos. Trans. R. Soc. Lond. B 358255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, J. W. A., M. L. Ginger, and S. J. Ferguson. 2004. Maturation of the unusual single-cysteine (XXXCH) mitochondrial c-type cytochromes found in trypanosomatids must occur through a novel biogenesis pathway. Biochem. J. 383537-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backman, K., M. Ptashne, and W. Gilbert. 1976. Construction of plasmids carrying cI gene of bacteriophage λ. Proc. Natl. Acad. Sci. USA 734174-4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bengtsson, J., C. Rivolta, L. Hederstedt, and D. Karamata. 1999. Bacillus subtilis contains two small c-type cytochromes with homologous heme domains but different types of membrane anchors. J. Biol. Chem. 27426179-26184. [DOI] [PubMed] [Google Scholar]
- 6.Bengtsson, J., H. Tjalsma, C. Rivolta, and L. Hederstedt. 1999. Subunit II of Bacillus subtilis cytochrome c oxidase is a lipoprotein. J. Bacteriol. 181685-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernard, D. G., S. Quevillon-Cheruel, S. Merchant, B. Guiard, and P. P. Hamel. 2005. Cyc2p, a membrane-bound flavoprotein involved in the maturation of mitochondrial c-type cytochromes. J. Biol. Chem. 28039852-39859. [DOI] [PubMed] [Google Scholar]
- 8.Colbert, C. L., Q. Wu, P. J. Erbel, K. H. Gardner, and J. Deisenhofer. 2006. Mechanism of substrate specificity in Bacillus subtilis ResA, a thioredoxin-like protein involved in cytochrome c maturation. Proc. Natl. Acad. Sci. USA 103410-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crow, A., R. M. Acheson, N. E. Le Brun, and A. Oubrie. 2004. Structural basis of redox-coupled protein substrate selection by the cytochrome c biosynthesis protein ResA. J. Biol. Chem. 27923654-23660. [DOI] [PubMed] [Google Scholar]
- 10.Crow, A., N. E. Le Brun, and A. Oubrie. 2005. The role of ResA in type II cytochrome c maturation. Biochem. Soc. Trans. 33149-151. [DOI] [PubMed] [Google Scholar]
- 11.Daltrop, O., J. W. A. Allen, A. C. Willis, and S. J. Ferguson. 2002. In vitro formation of a c-type cytochrome. Proc. Natl. Acad. Sci. USA 997872-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLano, W. L. 2002. The PyMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
- 13.Drygas, M. E., A. M. Lambowitz, and F. E. Nargang. 1989. Cloning and analysis of the Neurospora crassa gene for cytochrome c heme lyase. J. Biol. Chem. 26417897-17906. [PubMed] [Google Scholar]
- 14.Edeling, M. A., U. Ahuja, B. Heras, L. Thony-Meyer, and J. L. Martin. 2004. The acidic nature of the CcmG redox-active center is important for cytochrome c maturation in Escherichia coli. J. Bacteriol. 1864030-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erlendsson, L. S., R. M. Acheson, L. Hederstedt, and N. E. Le Brun. 2003. Bacillus subtilis ResA is a thiol-disulfide oxidoreductase involved in cytochrome c synthesis. J. Biol. Chem. 27817852-17858. [DOI] [PubMed] [Google Scholar]
- 16.Fabianek, R. A., H. Hennecke, and L. Thöny-Meyer. 1998. The active-site cysteines of the periplasmic thioredoxin-like protein CcmG of Escherichia coli are important but not essential for cytochrome c maturation in vivo. J. Bacteriol. 1801947-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feissner, R. E., C. L. Richard-Fogal, E. R. Frawley, J. A. Loughman, K. W. Earley, and R. G. Kranz. 2006. Recombinant cytochromes c biogenesis systems I and II and analysis of haem delivery pathways in Escherichia coli. Mol. Microbiol. 60563-577. [DOI] [PubMed] [Google Scholar]
- 18.Fortnagel, P., and E. Freese. 1968. Analysis of sporulation mutants. II. Mutants blocked in citric acid cycle. J. Bacteriol. 951431-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haima, P., S. Bron, and G. Venema. 1987. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol. Gen. Genet. 209335-342. [DOI] [PubMed] [Google Scholar]
- 20.Hederstedt, L. 1986. Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. Methods Enzymol. 126399-414. [DOI] [PubMed] [Google Scholar]
- 21.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204305-320. [DOI] [PubMed] [Google Scholar]
- 22.Hutchings, M. I., N. Shearer, S. Wastell, R. J. M. Van Spanning, and S. Spiro. 2000. Heterologous NNR-mediated nitric oxide signaling in Escherichia coli. J. Bacteriol. 1826434-6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadokura, H., H. P. Tian, T. Zander, J. C. A. Bardwell, and J. Beckwith. 2004. Snapshots of DsbA in action: detection of proteins in the process of oxidative folding. Science 303534-537. [DOI] [PubMed] [Google Scholar]
- 24.Kranz, R., R. Lill, B. Goldman, G. Bonnard, and S. Merchant. 1998. Molecular mechanisms of cytochrome c biogenesis: three distinct systems. Mol. Microbiol. 29383-396. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 26.Le Brun, N. E., J. Bengtsson, and L. Hederstedt. 2000. Genes required for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol. 36638-650. [DOI] [PubMed] [Google Scholar]
- 27.Lewin, A., A. Crow, A. Oubrie, and N. E. Le Brun. 2006. Molecular basis for specificity of the extracytoplasmic thioredoxin ResA. J. Biol. Chem. 28135467-35477. [DOI] [PubMed] [Google Scholar]
- 28.Marmur, J. 1961. Procedure for isolation of deoxyribonucleic acid from micro-organisms. J. Mol. Biol. 3208-218. [Google Scholar]
- 29.Moore, G. R., and G. W. Pettigrew. 1990. Cytochromes c: evolutionary, structural and physicochemical aspects. Springer-Verlag, Berlin, Germany.
- 30.Nicholas, K. B., and H. B. J. Nicholas. 1997. Genedoc: a tool for editing and annotating multiple sequence alignments. Distributed by the authors. http://www.nrbsc.org.
- 31.Ouyang, N., Y. G. Gao, H. Y. Hu, and Z. X. Xia. 2006. Crystal structures of E. coli CcmG and its mutants reveal key roles of the N-terminal beta-sheet and the fingerprint region. Protein Struct. Funct. Bioinform. 651021-1031. [DOI] [PubMed] [Google Scholar]
- 32.Page, M. D., Y. Sambongi, and S. J. Ferguson. 1998. Contrasting routes of c-type cytochrome assembly in mitochondria, chloroplasts and bacteria. Trends Biochem. Sci. 23103-108. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., F. E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 34.Schägger, H., and G. von Jagow. 1987. Tricine sodium dodecyl-sulfate polyacrylamide-gel electrophoresis for the separation of proteins in the range from 1 kDa to 100 kDa. Anal. Biochem. 166368-379. [DOI] [PubMed] [Google Scholar]
- 35.Schiött, T., and L. Hederstedt. 2000. Efficient spore synthesis in Bacillus subtilis depends on the CcdA protein. J. Bacteriol. 1822845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiött, T., M. Throne-Holst, and L. Hederstedt. 1997. Bacillus subtilis CcdA-defective mutants are blocked in a late step of cytochrome c biogenesis. J. Bacteriol. 1794523-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiött, T., C. von Wachenfeldt, and L. Hederstedt. 1997. Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis. J. Bacteriol. 1791962-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens, J. M., O. Daltrop, J. W. A. Allen, and S. J. Ferguson. 2004. C-type cytochrome formation: chemical and biological enigmas. Acc. Chem. Res. 37999-1007. [DOI] [PubMed] [Google Scholar]
- 39.Stirnimann, C. U., A. Rozhkova, U. Grauschopf, M. G. Grutter, R. Glockshuber, and G. Capitani. 2005. Structural basis and kinetics of DsbD-dependent cytochrome c maturation. Structure 13985-993. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 224673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thöny-Meyer, L., D. Stax, and H. Hennecke. 1989. An unusual gene-cluster for the cytochrome bc1 complex in Bradyrhizobium japonicum and its requirement for effective root nodule symbiosis. Cell 57683-697. [DOI] [PubMed] [Google Scholar]
- 42.Van der Oost, J., C. von Wachenfeldt, L. Hederstedt, and M. Saraste. 1991. Bacillus subtilis cytochrome oxidase mutants: biochemical analysis and genetic evidence for 2 aa3-type oxidases. Mol. Microbiol. 52063-2072. [DOI] [PubMed] [Google Scholar]
- 43.von Wachenfeldt, C., and L. Hederstedt. 1990. Bacillus subtilis 13 kDa cytochrome c550 encoded by cccA consists of a membrane anchor and a heme domain. J. Biol. Chem. 26513939-13948. [PubMed] [Google Scholar]
- 44.Yang, J., X. S. Liu, K. Bhalla, C. N. Kim, A. M. Ibrado, J. Y. Cai, T. I. Peng, D. P. Jones, and X. D. Wang. 1997. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 2751129-1132. [DOI] [PubMed] [Google Scholar]
- 45.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33103-119. [DOI] [PubMed] [Google Scholar]
- 46.Yu, J., and N. E. Le Brun. 1998. Studies of the cytochrome subunits of menaquinone:cytochrome c reductase (bc complex) of Bacillus subtilis. Evidence for the covalent attachment of heme to the cytochrome b subunit. J. Biol. Chem. 2738860-8866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.