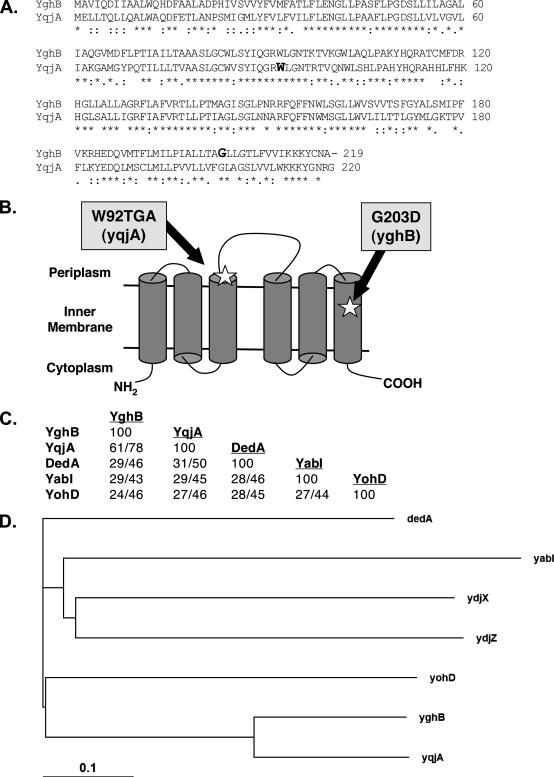
FIG. 1.
The E. coli DedA family of related predicted IM proteins. (A) Amino acid alignment between YqjA and YghB. W92 and G203, the sites of mutations in Lud135 in YqjA and YghB, respectively, are shown in bold. (B) Predicted IM topology of E. coli YghB and YqjA according to the Polyphobius prediction program (22) and experimental assignment of the C terminus to the cytoplasm (8). Both proteins are predicted to have six membrane spans and the indicated topology. Note that not all programs predict the same topology and alternate models are possible. The locations of the missense mutation in YghB (G203D) and the nonsense mutation in YqjA (W92TGA) in Lud135 are shown. (C) Percents identity and similarity between each DedA family member found in E. coli (45). All members display significant protein BLAST scores to each other (E values, <10−4), but there are no clearly conserved domains when all five proteins are aligned (not shown). YqjA and DedA display some similarity to YdjZ and YdjX, respectively (with 25 and 22% identity). However, other DedA family members are not significantly similar to YdjZ and YdjX, and so they are not included in this analysis for simplicity. (D) Phylogenetic tree showing evolutionary relationships between the E. coli proteins with similarity to DedA. The more distantly related YdjX and YdjZ were included for comparison. The analysis was performed using ClustalX (23).