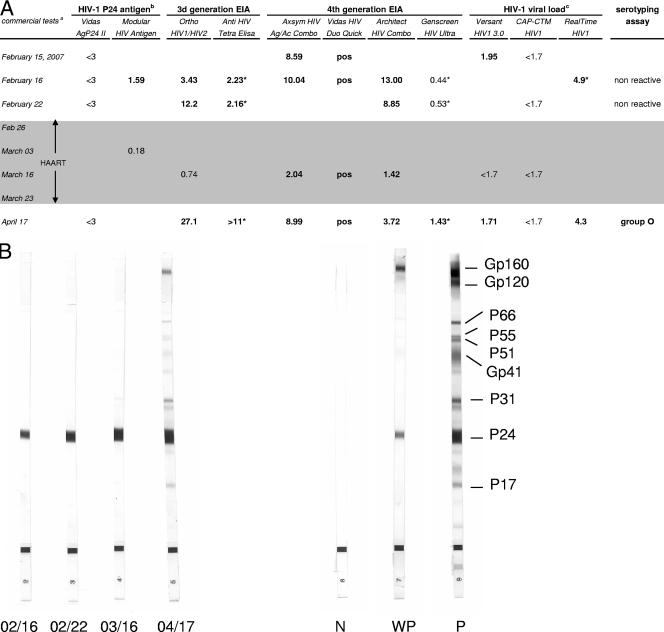
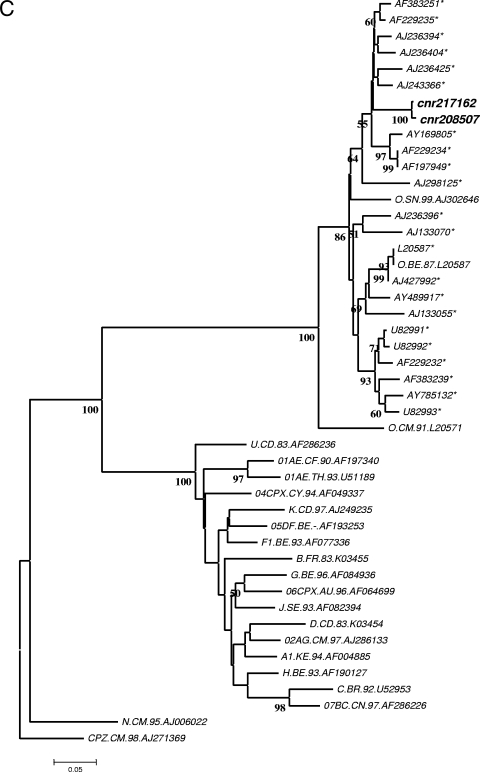
FIG. 1.
Serological and molecular findings on sequential samples from the patient reported. (A) Results of the different immunoassays and viral load assays. Bold indicates reactivity. Results of serological tests are expressed as signal-to-cutoff ratios. Signal-to-cutoff ratios of ≥1.00 were considered reactive, except for Vidas HIV Duo Quick (positive at >0.25). The shaded area indicates the period under HAART. (Superscript a) The commercial tests used were Vidas AgP24 II (bioMérieux), Modular HIV Antigen (Roche Diagnostics), Ortho HIV1/HIV2 Antibody Capture (Ortho Diagnostics), Anti-HIV Tetra ELISA (Biotest), Axsym HIV Ag/Ac Combo (Abbott), Vidas HIV Duo Quick (bioMérieux), Architect HIV Combo (Abbott), Genscreen HIV Ultra (Bio-Rad), Versant HIV-1 TNA 3.0 (Bayer), Cobas AmpliPrep-CobasTaqMan HIV-1 (CAP-CTM; Roche Diagnostics), and RealTime HIV-1 (Abbott Molecular). (Superscript b) HIV-1 p24 antigen levels are expressed in picograms per milliliter. (Superscript c) HIV-1 viral loads are expressed as log10 numbers of copies per milliliter. Asterisks indicate values obtained with samples that were analyzed retrospectively. (B) Western blot analysis of sequential serum samples from February to April 2007 (HIV-1/2 Blot; Genelabs). From left to right, the strips are shown in chronological order (month/day). N, negative control; WP, weakly positive control; P, positive control. (C) Phylogenetic tree. The env sequences (415 bp) from the index patient (cnr208507) and her partner (cnr2171262) were analyzed by the neighbor-joining method. The amplified sequences from the two patients were amplified and sequenced separately and compared with the 22 closest group O sequences (indicated by asterisks) selected from the Los Alamos database (http://hiv-web.lanl.gov) and 3 group O reference sequences and 17 group M reference sequences representative of various clades (one strain per pure subtype and major circulating recombinant form). Distances were calculated by the Kimura two-parameter method (T/t ratio = 2.0) and the MEGA analysis program (http://www.megasoftware.net). Bootstrap analysis was used to test the reliability of the branching order. Bootstrap values above 50 are indicated. The tree was rooted with a SIVcpz sequence (CPZ.CM.98.AJ271369) as the outgroup.