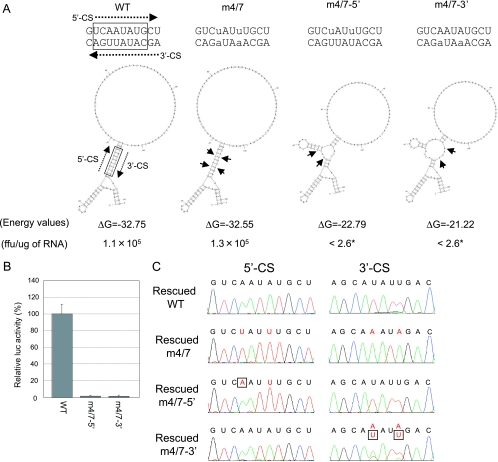
FIG. 5.
Effect of unmatched CS mutation. (A) Nucleotide sequence and secondary structures of the CS predicted by Mfold. Nucleotide substitutions at 5′ CS and 3′ CS are in lower case. Dotted arrows show the 5′-to-3′ direction of nucleotide in CS. Short arrows indicate nucleotide positions with substitutions. The ΔG values and the structures were produced using an artificial sequence created by fusing 52 bases of C coding region, 31 bases of 3′ UTR, and a 38-nucleotide A stuffer. The numbers of focus-forming units (ffu) per microgram of electroporated RNA are also indicated. Asterisks show no detection of foci. The limit of detection for this assay was 2.6 focus-forming units/μg. (B) RNA replication of unmatched CS mutant replicons in BHK cells. Average triplicate relative luciferase activities at 2 days posttransfection were normalized to those at 5 h, and the WT control value was set to 100%; error bars show standard deviations. The experiment was repeated at least twice, with consistent results. (C) Electropherograms showing the sequences of the blind-passed replicons derived from WT, m4/7, m4/7-5′, and m4/7-3′. The nucleotide sequences of the CS are indicated above the electropherograms. Deliberately mutated CS nucleotides are shown in red; changes from these mutations detected in rescued populations are shown in red and boxed in black.