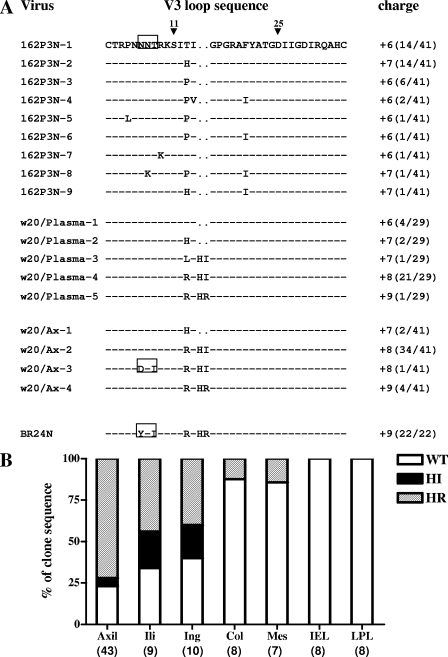
FIG. 1.
(A) Comparison of V3 loop sequences of the parental R5 SHIVSF162P3N isolate, the recovered X4 virus SHIVBR24N, and viruses present in plasma and axillary (Ax) LN of macaque BR24 at 20 wpi. Dashes denote similarity in sequences, and gaps are indicated as dots. The net positive charge of this region is indicated in the right column. Positions 11 and 25 within the V3 loop that frequently distinguished HIV-1 X4 and R5 viruses (16) are designated by arrowheads, and the highly conserved PNG site at the V3 base is boxed. The numbers in parentheses represent the number of clones with the indicated V3 loop sequence reported relative to the total number of clones sequenced. (B) Representation of SHIV viral variants without (WT) or with HI or HR insertions in various tissue sites of macaque BR24 at the time of death (28 wpi). Percentages of Env clones present in axillary (axil), iliac (ili), inguinal (ing), colonic (col), and mesenteric (mes) LNs, as well as intraepithelial lymphocytes (IEL) and LPL of the gut with the indicated signature V3 loop sequences are shown. Numbers in parenthesis indicate the number of gp120 clones sequenced from each of the tissue sites.