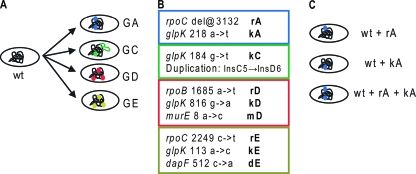
FIG. 1.
Origin of the strains utilized in the present study. (A) Parallel adaptive evolutions produced the four endpoint strains analyzed. (B) The mutations acquired by the endpoint strains were identified by whole-genome resequencing and are listed in the colored boxes. The short-hand notation for each mutation indicates the effected gene (r = rpoB/C, k = glpK, m = murE, d = dapF) and the endpoint strain it was found in (i.e., kA = glpK mutation identified in strain GA) (9). (C) Allele replacement strains were then constructed using gene gorging (8, 9) to introduce the mutations to the wild-type genetic background. An allele replacement strain was created that carries each mutation identified in isolation, as well as strains that carry multiple or all mutations identified in an endpoint strain.