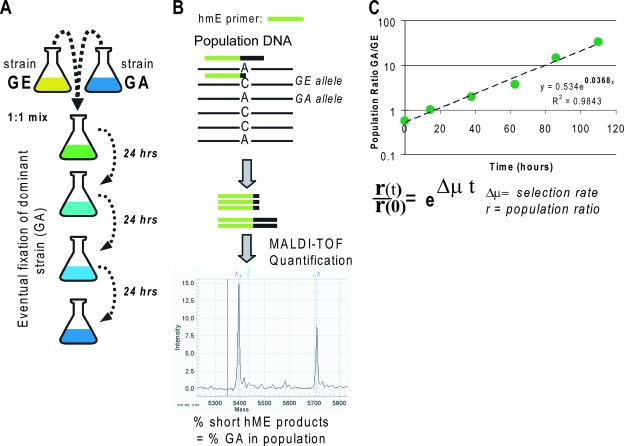
FIG. 2.
Design of the competition experiments. (A) The two strains to be competed are inoculated into a single vessel to compete directly for media resources. The culture is propagated for 3 days by serial passage, with samples collected daily to determine the current population frequency. (B) The population frequency at each time point is based on the allele frequency of the known mutations in each strain. DNA isolated daily from the competing culture is subjected to heterogeneous mass-extension assays (Sequenom, Inc., San Diego, CA) to determine the frequency of the mutations that distinguish the competing strains. The 200-bp region around each mutation is PCR amplified (results not shown), followed by PCR extension of an hME primer that binds directly upstream of the mutation. Extension of the primer is performed with nucleoside triphosphate mix missing a species of nucleotide that complements one of the alleles, causing amplification of mutation-harboring alleles to terminate at different loci than the wild type, producing products with distinct masses. All of the primers are listed in Table S2 in the supplemental material. The relative amounts of the extension products are determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry from the ratio of the product peak areas. (C) We calculate the estimated selection rate as the Malthusian parameter of the change in the population ratio over time (14). Assay bias was negligible in most cases (see Table S3 in the supplemental material).