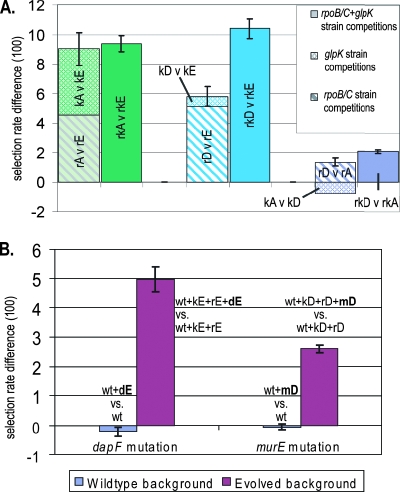
FIG. 4.
Epistatic interactions suggested by additive analysis. Epistatic interactions can be inferred when mutations cause different selection rate increases in different genetic backgrounds. (A) Each pair of bars shows the selection rates from competitions between mutations from the same endpoint strains. The selection rates represented in the left column result from competitions between allele replacement strains harboring single mutations; the horizontally striped bar represents the selection rate difference between the rpoB/C mutations of two endpoint strains (Fig. 3B), and the cross-hatched bar represents the selection rate difference between the glpK mutations from the same two endpoint strains (Fig. 3C). The solid-colored right-hand bar represents the selection rate difference found from competitions between allele replacement strains that carried both of a given endpoint strain's glpK and rpoB/C mutations (Fig. 3D). Significant discrepancies between the heights of each pair of bars indicate epistatic interactions between the glpK and rpoB/C mutations of one or both of the involved endpoint strains. An asterisk indicates the value does not agree well with estimates extrapolated from the rest of the set and may be questionable (see Discussion). (B) The adaptive values of the murE and dapF mutations increase in the evolved genetic background relative to wild type. Competitions were performed in both the wild-type and the evolved genetic background between strains that differed only by the murE or dapF mutation. Both mutations had effectively no adaptive value in the wild-type genetic background but were clearly adaptive in the presence of their coacquired rpoB/C and glpK mutations.