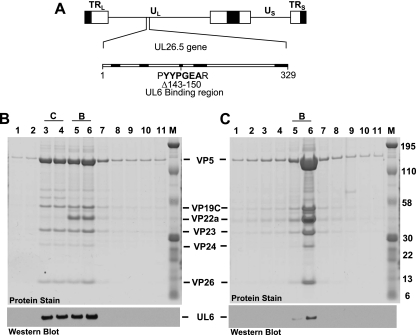
FIG. 1.
(A) Physical map of the HSV-1 genome showing the location of the UL26.5 gene, which encodes the 329-amino-acid VP22a scaffold protein. UL and US, long and short unique region sequences. The next line shows the location of the eight amino acids (amino acids 143 to 150) deleted from the scaffold mutant (bvFH411) that previously were shown to be essential for interaction with the UL6 portal protein. Regions that are conserved in the scaffold proteins of alphaherpesviruses are in bold. (B and C) Nuclear lysates of Vero cells infected at an MOI of 5 with KOS (B) or bvFH411 (C) were subjected to sucrose gradient sedimentation, and the protein composition for each gradient fraction was determined by SDS-PAGE (stained gel). Lane 1 is the bottom and lane 11 is the top of the gradient. Gradient fractions that contained C and B capsids are indicated (there was no A capsid band present in KOS gradient). The positions of the capsid proteins (major capsid protein, VP5; triplex proteins, VP19C and VP23; scaffold protein, VP22a; protease, VP24; smallest capsid protein, VP26) are indicated. Molecular mass standards are visible in lane M (kDa). In the bottom panels, gradient fractions were analyzed by SDS-PAGE followed by immunoblotting for UL6 using the 1C9 antibody (16).