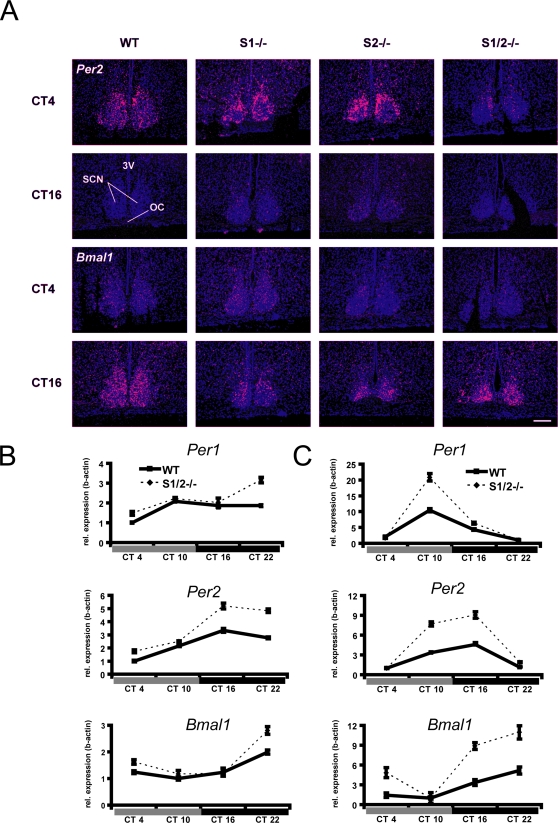
Figure 3. Altered clock gene expression in the SCN, cortex and liver.
(a) In situ hybridization at the level of the SCN using 35S-labeled riboprobes directed against Per2 and Bmal1 mRNAs. In WT, S1−/− and S2−/−, Per2 mRNA expression in the SCN is strongly elevated at CT4 compared to CT16 reflecting a clock-controlled rhythm of expression. In S1/2−/− mice, the increase of Per2 mRNAs expression during the subjective day is blunted. Bmal1 expression is elevated at CT16 in all genotypes (3V, third ventricle; SCN, suprachiasmatic nucleus; OC, optic chiasm). Scale bar = 200 µm. (b,c) Relative gene expression changes during four consecutive circadian timepoints (CT4, 10, 16 and 22) in wild-type (WT) and Sharp-1/-2 double mutant (S1/2−/−) cerebral cortex (b) and liver (c) revealed by qRT-PCR. The amplitude of the circadian regulated gene expression of Per1, Per2 and Bmal1 is significantly enhanced during the first day of DD comparing WT and S1/2−/− mice (p<0.05 for Per1 in the cortex, all other comparisons p<0.01 with 2-way ANOVA and the factors ‘genotypes×time’). (Data represent mean values ±SD, n = 3 per genotype).