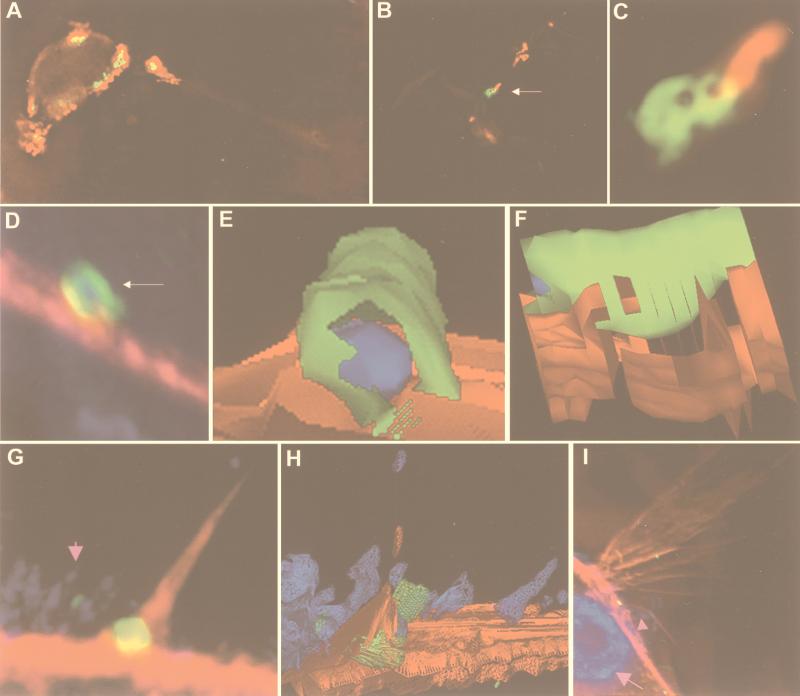
Figure 3.
Deconvolution immunofluorescence and 3D modeling of H. pylori attached to AGS cells. H. pylori strain G27 (A–F), strain 87A300 (G–I). (A) H. pylori attached to AGS cells and stained for H. pylori (red) and CagA (green). Colocalization is yellow. The field is a 0.2-μm image. (B) H. pylori attached to AGS cells and stained for H. pylori (red) and CagA (green). Colocalization is yellow. The field is a volume composite of 0.2-μm images. (C) The region indicated by the arrow in B is magnified in C. (D) H. pylori attached to AGS cells and stained for H. pylori (blue), CagA (green), and actin (red). Colocalization of green and red appears yellow, of blue and green, cyan, and of red and blue, magenta. The field is a volume composite of 0.2-μm images. The region indicated by the arrow in D was used to generate the 3D solid models shown in E and F. F is a side view of E. (E and F) H. pylori is blue, CagA is green, and actin is red. (G) H. pylori (small pink arrow) attached to AGS cells and stained for H. pylori (blue), CagA (green), and actin (red). Colocalization of green and red appears yellow, of blue and green, cyan, and of red and blue, magenta. The field is a 0.2-μm image. The field in G was used to generate the 3D wire model shown in H. (H) H. pylori is blue, CagA is green, and actin is red. (I) H. pylori (small pink arrow) attached to AGS cells were stained with DAPI [which stains the host-cell nucleus (large pink arrow) and bacterial nuclear DNA blue], anti-CagA (green), and actin (red). Colocalization of green and red appears yellow, of blue and green, cyan, and of red and blue, magenta. The field is a volume composite of 0.2-μm images. Imaged and modeled on an Applied Precision DeltaVision Deconvolution System.