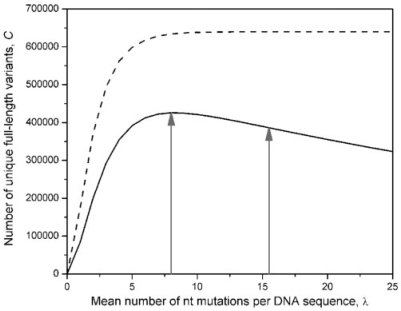
Figure 1.
The estimated numbers of unique DNA sequence variants (CDNA, dashed line) and protein sequence variants (Cprotein, solid line) in a purF epPCR library (24), plotted as a function of the DNA mutation rate, λ. The epPCR comprised 30 thermal cycles, with eff = 0.41. The library contained 6.4 × 105 clones. A total of 7549 bp of DNA sequence was obtained from randomly chosen library members, and 77 substitutions plus one single-nucleotide deletion were observed. These data were used to construct the nucleotide mutation matrix for PEDEL-AA. The library used for genetic selection experiments contained λ = 15.5 mutations per sequence; the estimated sequence diversity it contains is indicated by the vertical arrow on the right. The maximally diverse library is indicated by the vertical arrow at λ = 8. Note that, after peaking, the number of unique amino acid (but not nucleotide) variants decreases with increasing λ, due to an increasing number of truncated sequences.