Abstract
MAGNOLIA is a new software for multiple alignment of nucleic acid sequences, which are recognized to be hard to align. The idea is that the multiple alignment process should be improved by taking into account the putative function of the sequences. In this perspective, MAGNOLIA is especially designed for sequences that are intended to be either protein-coding or structural RNAs. It extracts information from the similarities and differences in the data, and searches for a specific evolutionary pattern between sequences before aligning them. The alignment step then incorporates this information to achieve higher accuracy. The website is available at http://bioinfo.lifl.fr/magnolia.
INTRODUCTION
More and more newly sequenced genomes are becoming available every week. Tiling arrays are also gaining popularity for detecting novel transcripts in sequenced genomes. In this context, sequence annotation is an essential step in understanding the genome and the transcriptome of a species. Comparative genomics has proven to be a promising approach to address this problem. Large-scale comparisons of prokaryotic and eukaryotic genomes reveal thousands of conserved regions obtained by homology or synteny. These regions might be protein-coding sequences (1) or non-coding RNA genes (2,3,4). Annotation by comparative analysis typically involves two steps: first aligning the sequences, then analysing the multiple alignment to detect an evolutionary pattern that is representative of the selection pressure. This idea is exploited in Exoniphy (5) for exon detection, in RNAz (6) or Evofold (7) for structural RNA prediction or in Qrna (8), that implements both a coding and an non-coding model for RNAs. In this computational protocol, high-quality sequence alignment is an essential prerequisite step. This task, however, is difficult because sequence similarity is often reduced at the nucleic level. Regarding protein coding genes, nucleic acid sequences exhibit a much larger sequence heterogeneity compared to their encoded amino acid sequences due to the redundancy of the genetic code. It is well known that the combination of nucleic acid and amino acid sequence information leads to improved alignments (9,10). The same situation holds for non-coding RNA genes. The spatial structure evolves slower than its primary structure. So pure-sequence-based multiple alignment tools perform poorly on low-homology datasets of structural RNAs (11). In this article, we present the MAGNOLIA website, whose objective is to provide an advanced tool for aligning nucleic acid sequences. The idea is to get rid of the dichotomy between aligning and predicting the function. If we assume that sequences are either protein-coding or structural RNAs, it is possible to incorporate some functional information into the alignment algorithm to improve the result. The multiple alignment can then be used as a starting point to refine the comparative analysis or to carry out further predictions, such as motif finding or phylogeny reconstruction.
METHODS
The method has two steps. First, it tries to predict the function of the sequences according to the substitution pattern between sequences. Second, a multiple alignment is built based on the putative function of the sequences. If the sequences are recognized as coding sequences, then the multiple alignment uses the amino acid sequences. If the sequences are recognized to contain a conserved secondary structure, then the mutiple alignment takes into consideration long-range base pair interactions.
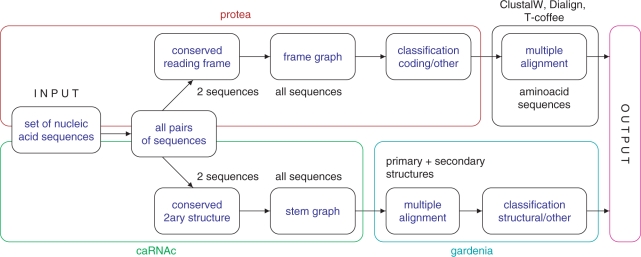
MAGNOLIA includes three specific modules: Protea for protein coding sequences, caRNAc and gardenia for structural RNAs. The overall scenario is summed up in Figure 1.
Figure 1.
MAGNOLIA scenario.
Protea implements an evolutionary model for protein-coding sequences (12). Here the idea is that the selection pressure tends to preserve the encoded amino acid sequence, and it is possible to identifies coding sequences by looking for a global conservation of common reading frames. The method first identifies best potential reading frames from each pair of sequences, and then incorporates this information into a frame graph from which a coding significancy score is calculated. By doing so, it also predicts the associated reading frame for each sequence. If the sequences are classified as protein-coding sequences, then the multiple alignment of nucleic acid sequences is built from the hypothetical amino acid sequences using ClustalW (13), Dialign2 (10) or T-coffee (14). caRNAc is for structural RNA genes (15). In this model, the selection pressure tends to preserve the secondary structure of the molecule, and mutations should retain the ability to form base pairs into energetically favorable stems.
caRNAc is able to recover a conserved secondary structure from a set of unaligned sequences. This idea is also present in refs. (16,17), that fold and align several sequences at the same time, for example. But these programs are still computationally demanding. We circumvent the problem by using a heuristics approach. The algorithm uses a Sankoff-based dynamic programing approach to identify conserved strutures for all pairs of sequences. Then all pairwise foldings are combined into a graph-theoretical structure called the stem graph. Only frequent common stems that correspond to highly connected subgraphs in the stem graph are retained.
Gardenia is used to build the multiple alignment for potential structural RNA sequences. The method takes into account both the nucleic sequence and the putative common secondary structure predicted by caRNAc. It relies on the dynamic programming algorithm for pairwise comparison proposed in ref. (18). RNA sequences are encoded as arc-annotated sequences, and a multiple alignment for a set of arc-annotated sequences is a nested common supersequence. The edit scheme incorporates evolutionary operations concerning free bases (base substitution, base deletion) and base pairs (arc-mismatch, arc-removing, arc-breaking, arc-altering), originally defined in ref. (19). It is easy to show that this problem is NP-hard. We take a heuristic approach and use a progressive method. The method starts with constructing all pairwise alignments to determine the degree of similarity for each pair of sequences. Then it combines sequences into a multiple alignment by an ascending hierarchical clustering. Pairwise alignment of supersequences rely on the same algorithm as pairwise alignments for arc-annotated sequences. This is made possible because supersequences can be viewed as a nested arc-annotated sequences on an extended alphabet. The score of one node is its SP (sum-of-pairs) score. Lastly, the space search of the dynamic programming alignment is pruned using constraints coming from the caRNAc output. This provides a significant speed up.
WEB SERVER
Input
MAGNOLIA requires as input data a set of RNA or DNA sequences in the standard FASTA format. This set should contain at least two distinct sequences and at most ten sequences. It can be stored in a file to be uploaded to the server, or pasted directly in the text box.
Output
A typical run of MAGNOLIA takes a few seconds. Upon completion of a job, MAGNOLIA displays the result on a new web page. The job assigned a unique identifier that can be used to retrieve results for one week. All results are available for download in Clustal and bracket-dot format.
If input sequences are annotated as coding sequences, then two multiple alignments are displayed. The first alignment is built on the putative amino acid sequences obtained by virtual translation using the predicted reading frame, and the second alignment is the corresponding alignment on nucleic acid sequences obtained by reverse translation, allowing for frameshifts. Codons in the nucleic acid sequences are put in color: two base triplets coding for the same amino acid bear the same color. The color choice is inspired from the RasMol amino acid color scheme. Figure 2 shows an example of MAGNOLIA output obtained with a family of protein-coding sequences.
Figure 2.
Alignment for Zn-finger in Ran binding proteins (PFAM PF00641). The average length is 92 nt and the average identity percentage is 45.1%. Triplets are colorized according to the encoded amino acid. The reference alignment provided in Pandit is almost identical (20).
If input sequences are annotated as structural RNA genes, then a multiple alignment taking into consideration the primary structure accompanied by the secondary structure is displayed. Concerning the secondary structure, base pairings are indicated in bracket-dot format: Each base-pair is represented by a pair of matching brackets and unpaired bases are represented by dots. The lack of pseudoknots in the secondary structure ensures that this notation defines a unique folding. Moreover, stems in the alignment are highlighted in colors. Figure 3 shows an example of output obtained with a family of non-coding RNAs. For each sequence, the individual putative secondary structure is also provided in five formats: CT, JPEG, PS, bracket-dot format and as a list of constrained base pairings. JPEG and PS files are automatically produced from the CT file using the NAview layout program (22).
Figure 3.
Alignment for five tRNA sequences (BRaliBase II – aln 13). The average length is 76 nt and the average identity percentage is 51.0%. MAGNOLIA finds three common stems (in blue, green and orange). All pairings are correct, and the multiple alignment is consistent with the reference alignment available in RFAM-RF00005 (21).
Some data sets are not identified as coding RNAs nor as non-coding RNAs. The first possibility is that the sequences might have an alternative function, such as untranslated regions in messenger RNAs, promoter elements, etc. The second possibility is that the sequences are highly conserved. In this context, the comparative analysis approach used by MAGNOLIA is not suitable. The evolutionary signal is too weak and the sequences do not exhibit any significant mutational bias towards any model. This is an intrinsic limitation of the method. But this limitation is harmless for practical purposes, because standard multiple sequence alignment tools usually yield good results on high-identity data sets. So when the average identity percentage is greater than 90%, the server outputs a warning message and provides a default multiple alignment constructed directly with ClustalW on the initial data set.
One final point worth mentioning is that the classification is not mutually exclusive. Some sequences might contain conserved secondary structure elements within a coding region. Two such examples are the cis-acting regulatory element from the human rhinoviruses, that is located in the open reading frame of the capsid proteins [RFAM – RF00220 (21)], or the Hepatitis C stem-loop VII structure found in the coding region of the RNA-dependent RNA polymerase gene NS5B [RFAM – RF000468 (21)]. In such cases, MAGNOLIA releases two multiple alignments.
EXPERIMENTAL RESULTS
We evaluate the accuracy of the method on two large data sets: Pandit (20) and BRaliBase II (11). Pandit is a registry of families of homologous protein domains, accompanied by curated RNA sequence alignments. BRaliBase II is a set of non-coding RNA families that has been used to establish a benchmark of multiple sequence alignment programs upon structural RNAs. It is composed of four families: Group II introns, 5S rRNA, tRNA and U5 spliceosomal RNA.
Results on pandit database
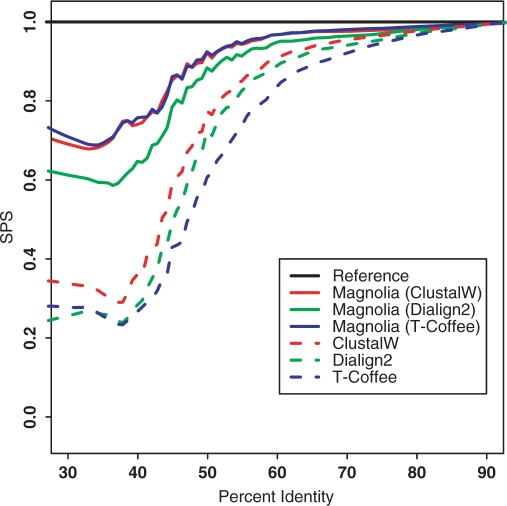
For each family, we selected a subset of four sequences at random. It remains 6491 families, whose average sequence length is 604 bp. 6122 (94.3%) families are correctly classified as coding sequences, among them more than 99% with the correct reading frame predicted for each sequence. Less than 3% of the families are classified as structural RNA. To estimate the quality of the alignments, we used the sum-of-pairs score (SPS) of the Baliscore software (23). The SPS is calculated such that it increases with the number of sequences correctly aligned. We compared MAGNOLIA with ClustalW, T-coffee and Dialign2 on the same nucleic acid sequences. Results are displayed in Figure 4.
Figure 4.
MAGNOLIA alignments on Pandit. The x-axis represents the average identity percentage and the y-axis represents the SPS value. For MAGNOLIA, we tried all possible combinations concerning the alignment tool in the amino acid alignment step: ClustalW, T-coffee and Dialign2. For Dialign2, we selected the appropriate option Translation of nucleotide diagonals into peptide diagonals when comparing the nucleic acid sequences. MAGNOLIA clearly outperforms other methods.
Results on BRaliBase II benchmark data
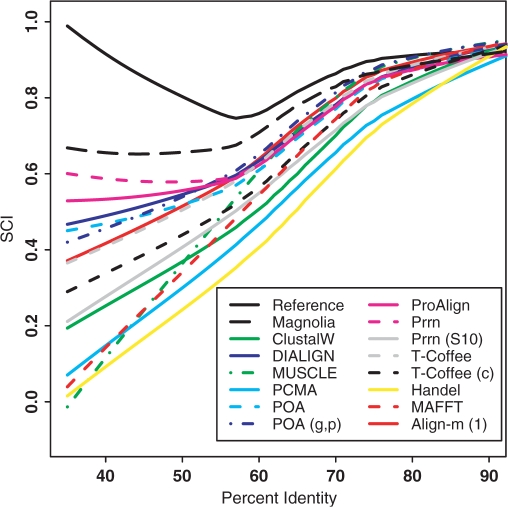
This benchmark contains 388 alignments, that are classified into high, medium and low identity data sets. MAGNOLIA failed to identify a structural evolutionary pattern for 20% of them and falsely assigned a protein coding function for 7% of them. Following ref. (11), we use the structure conservation index (SCI) to assess the accuracy of alignments. This score provides a measure of the conserved secondary structure information contained within the alignment. Results for MAGNOLIA are reported in Figure 5, together with results for other alignment tools used in the benchmark. We also evaluated the accuracy of the secondary structure found by MAGNOLIA and compared it to two recent structural alignment programs: Murlet (16) and Mlocarna (17). For each software and for each identity class, we computed the percentage of correct base pairings amongst the set of predicted base pairings. Results are shown in Table 1. It appears that MAGNOLIA has similar performances as Murlet and outperforms Mlocarna. Furthermore, the total runtime is more than 12 times faster for MAGNOLIA than for the two other methods (< 20 min for the whole data set, compared to more than 4 hours).
Figure 5.
MAGNOLIA alignments on BRaliBase II. The x-axis represents the average identity percentage and the y-axis represents the SCI value. The above curve is calculated from reference alignments. MAGNOLIA appears to be the closest curve to the reference curve for identity percentage ranging from 40% to 80%. For higher identity percentages, all methods show similar performances.
Table 1.
Accuracy percentage for MAGNOLIA, Murlet and Mlocarna for reference secondary structures of BRaliBase II
| Identity class | |||
|---|---|---|---|
| Low | Medium | High | |
| MAGNOLIA | 72.0% | 76.3% | 87.0% |
| Murlet | 78.1% | 76.2% | 77.8% |
| Mlocarna | 68% | 71.1% | 78.9% |
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by CNRS (Centre National de la Recherche Scientifique).
Conflict of interest statement. None declared.
REFERENCES
- 1.Zhu J, Sanborn JZ, Diekhans M, Lowe CB, Pringle TH, Haussler D. Comparative genomics search for losses of long-established genes on the human lineage. PLoS Comput. Biol. 2007;3 doi: 10.1371/journal.pcbi.0030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Washietl S, Hofacker IL, Lukasser M, Hut-tenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 3.Torarinsson E, Yao Z, Wiklund ED, Bramsen JB, Hansen C, Kjems J, Tommerup N, Ruzzo W, Gorodkin J. Genome Res. 2007. Comparative genomics beyond sequence-based alignments: RNA structures in the ENCODE regions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Washietl S, Pedersen JS, Korbel JO, Stocsits C, Gruber AR, Hackermüller J, Hertel J, Linde-meyer M, Reiche K, et al. Structured RNAs in the ENCODE selected regions of the human genome. Genome Res. 2007;17:852–864. doi: 10.1101/gr.5650707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siepel A, Haussler D. Research in computational molecular biology (RECOMB) New York, NY, USA: ACM Press; 2004. Computational identification of evolutionarily conserved exons. pp. 177–186. [Google Scholar]
- 6.Washietl S, Hofacker IL, Stadler PF. Fast and reliable prediction of noncoding RNAs. Proc. Natl Acad. Sci. USA. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen JS, Bejerano G, Siepel A, Rosen-bloom K, Lindblad-Toh K, Lander E, Rogers J, Kent J, Miller W, Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2000;2 doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivas E, Eddy SR. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics. 2001;2:8. doi: 10.1186/1471-2105-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stocsits RR, Hofacker I, Fried C, Stadler P. BMC Bioinformatics. 2005. Multiple sequence alignments of partially coding nucleic acid sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgenstern B. Dialign 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics. 1999;15:211–218. doi: 10.1093/bioinformatics/15.3.211. [DOI] [PubMed] [Google Scholar]
- 11.Gardner PP, Wilm A, Washietl S. A benchmark of multiple sequence alignment programs upon structural RNAs. Nucleic Acids Res. 2005;33:2433–2439. doi: 10.1093/nar/gki541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontaine A, Touzet H. Bioinformatics and Biomedecine (BIBM) San Franscisco, USA: IEEE Computer Society; 2007. Computational identification of protein-coding sequences by comparative analysis. pp. 95–102. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 15.Touzet H, Perriquet O. CARNAC: folding families of related RNAs. Nucleic Acids Res. 2004;W32:142–145. doi: 10.1093/nar/gkh415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiryu H, Tabei Y, Kin T, Asai K. Murlet: a practical multiple alignment tool for structural RNA sequences. Bioinformatics. 2007;23:1588–1598. doi: 10.1093/bioinformatics/btm146. [DOI] [PubMed] [Google Scholar]
- 17.Will S, Reiche K, Hofacker IL, Stadler PF, Backofen R. Inferring non-coding RNA families and classes by means of genome-scale structure-based clustering. PLOS Comput. Biol. 2007;3 doi: 10.1371/journal.pcbi.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blin G, Touzet H. String Processing and Information Retrieval (SPIRE) 2006. How to compare arc-annotated sequences: the alignment hierarchy. pp. 291–303. Vol. 4209 of Lecture Notes in Computer Science. [Google Scholar]
- 19.Jiang T, Lin G, Ma B, Zhang K. A general edit distance between RNA structures. J. Comput. Biol. 2002;9:371–388. doi: 10.1089/10665270252935511. [DOI] [PubMed] [Google Scholar]
- 20.Whelan S, de Bakker P, Quevillon E, Rodriguez N, Goldman N. PANDIT: an evolution-centric database of protein and associated nucleotide domains with inferred trees. Nucleic Acids Res. 2006;34:D327–D331. doi: 10.1093/nar/gkj087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. RFAM: an RNA family database. Nucleic Acids Res. 2003;33:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruccoleri RE, Heinrich G. An improved algorithm for nucleic acid secondary structure display. Comput. Appl. Biosci. 1988;4:167–173. doi: 10.1093/bioinformatics/4.1.167. [DOI] [PubMed] [Google Scholar]
- 23.Thompson JD, Plewniak F, Poch O. BA1-iBASE: a benchmark alignment database for the evaluation of multiple alignment programs. Bioinformatics. 1999;15:87–88. doi: 10.1093/bioinformatics/15.1.87. [DOI] [PubMed] [Google Scholar]