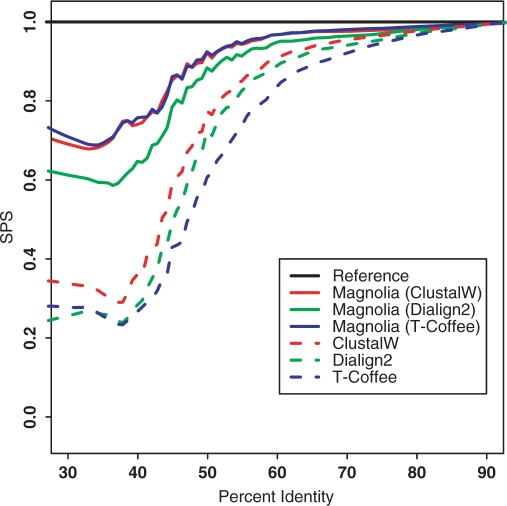
Figure 4.
MAGNOLIA alignments on Pandit. The x-axis represents the average identity percentage and the y-axis represents the SPS value. For MAGNOLIA, we tried all possible combinations concerning the alignment tool in the amino acid alignment step: ClustalW, T-coffee and Dialign2. For Dialign2, we selected the appropriate option Translation of nucleotide diagonals into peptide diagonals when comparing the nucleic acid sequences. MAGNOLIA clearly outperforms other methods.