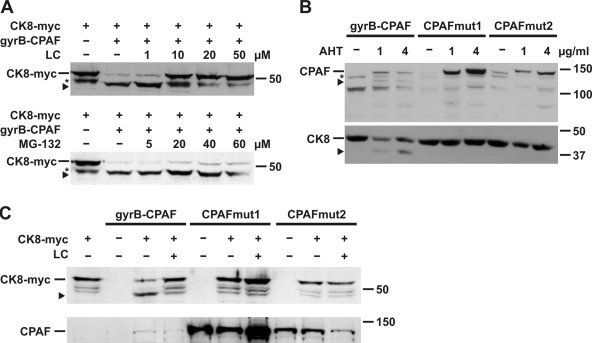
Figure 2.
Analysis of proteolytic activity of CPAF. (A) Inhibition of CPAF activity by proteasome inhibitors. Cell extracts of CPAF K6 cells (gyrB-CPAF) or T-REx-293 cells expressing myc-tagged cytokeratin 8 (CK8-myc) were combined in the presence of various amounts of the proteasome inhibitors lactacystin (LC; top) or MG-132 (bottom) as indicated, and analyzed by Western blotting using an antibody specific for the myc tag. The arrowhead indicates a cleavage product of CK8-myc (asterisk marks an unspecific signal). (B) Analysis of the proteolytic activity of CPAF mutants in vivo. T-REx-293 cells were transfected with either the gyrB-CPAF construct or one of two CPAF mutants. In CPAFmut1, the Tsp-active site was mutated by the replacement S491A. In mutant 2 (CPAFmut2), two amino acid residues (L273G, S275V) were exchanged to prevent autocatalytic cleavage of CPAF. CPAF expression was induced by AHT as indicated. Cell extracts were analyzed by Western blotting using either FLAG tag (top) or CK8 antibodies (bottom). The arrowhead in the top panel shows the cleavage product very likely corresponding to gyrB-CPAF-N; the arrowhead in the bottom panel indicates a cleavage product of CK8. An unspecific signal is marked by an asterisk. (C) Analysis of the proteolytic activity of CPAF mutants in vitro. T-REx-293 cells were transfected with the CPAF constructs as indicated, and cell lysates were incubated with extracts containing myc-tagged CK8, and analyzed as described in A (top). Expression of the CPAF constructs was confirmed by Western blotting using a FLAG tag antibody (bottom).