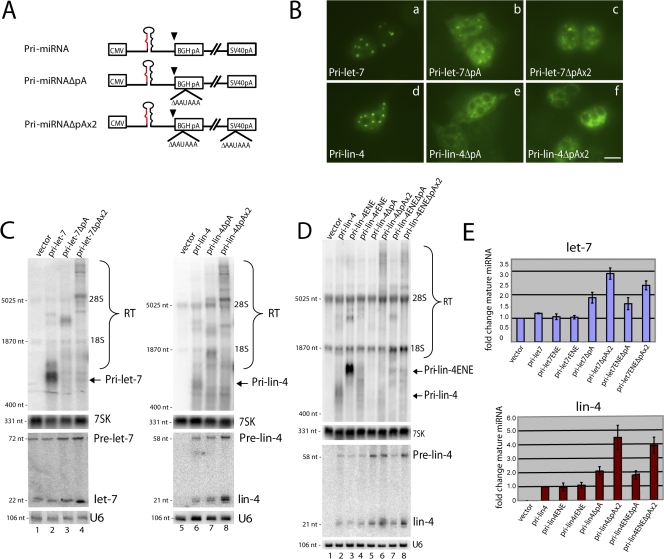
Figure 2.
Pri-miRNAs lacking a CPA signal do not localize to nuclear foci and are processed with increased efficiency. (A) Pri-miRNA constructs are diagramed as in Fig. 1 A; arrowheads indicate the location of five copies of the ENE in the forward or reverse complement (r) orientation. Deleted vector-encoded CPA signals are indicated by ΔAAUAAA. (B) ISH performed on cells transfected with constructs diagramed in A. Bar, 10 μm. (C and D) Northern blot analysis of RNA from cells transfected as in B. Note that transfections in C (lanes 6–8) are repeated in D (lanes 2, 5, and 6) to allow comparison of miRNA levels. Cross-hybridization with 18S and 28S ribosomal RNA is indicated. RT, readthrough transcripts. (E) Comparison of mature miRNA levels produced from the constructs in A, quantified from Northern blots similar to those shown in C and D. miRNA levels were normalized to U6 snRNA; levels of endogenous let-7 (top) or levels of lin-4 from pri-lin-4 transfections (bottom) were set to 1. Error bars indicate standard deviations (pri-let-7, n = 3; pri-lin-4, n = 5).