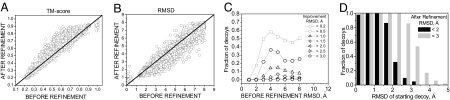
Fig. 1.
For all refinement trajectories, results of decoy structure refinement within each trajectory (100 decoys per protein, 39 proteins). (A and B) TM-score (A) and Cα rmsd (B) to the native structure of the lowest-energy decoy after refinement versus decoy's initial TM-score (A) or Cα rmsd (B). (C) Fraction of decoys that refined by more than a given Cα rmsd threshold (for 0.2-, 0.5-, 1.0-, 1.5-, and 2.0-Å rmsd thresholds) with respect to their initial native similarity. (D) Fraction of decoys that refined to (or remained within) the accuracy of 2-Å (black) and 3-Å (gray) Cα rmsd to the native state with respect to their initial native similarity. Fractions of decoys were calculated in 1-Å bins.