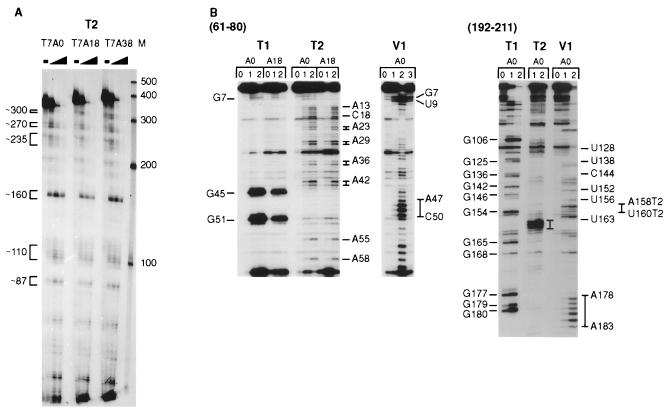
Figure 1.
Enzymatic probing analysis of T7A0, T7A18, and T7A38 MFA2 RNAs. (A) The RNAs were labeled at the 5′ end and cleaved with RNase T2 as described in Materials and Methods. The amount of RNase T2 used for each RNA was none, 0.002, 0.006, and 0.02 unit, and a 6% polyacrylamide gel was used for analysis. Ambion (Austin, TX) RNA markers are shown on the right. (B) RNAs were cleaved with RNase T1, T2, and V1. For RNase T1, the lanes labeled 0, 1, and 2 were no, 0.04, and 0.08 unit of enzyme. For RNase T2, the same lanes were no, 0.0012, and 0.004 unit. The RNase V1 cleavage reactions were done with no (0), 0.04 (1), 0.2 (2), and 1.2 (3) unit of enzyme. Primer extension analysis was done as described under Materials and Methods using primers complementary to the MFA2 sequence shown at the top. The T1 cleavage sites are shown on the left and the T2 and V1 sites on the right. With the nucleotide 192–211 antisense primer, the T2 sites are designated T2 after the site.