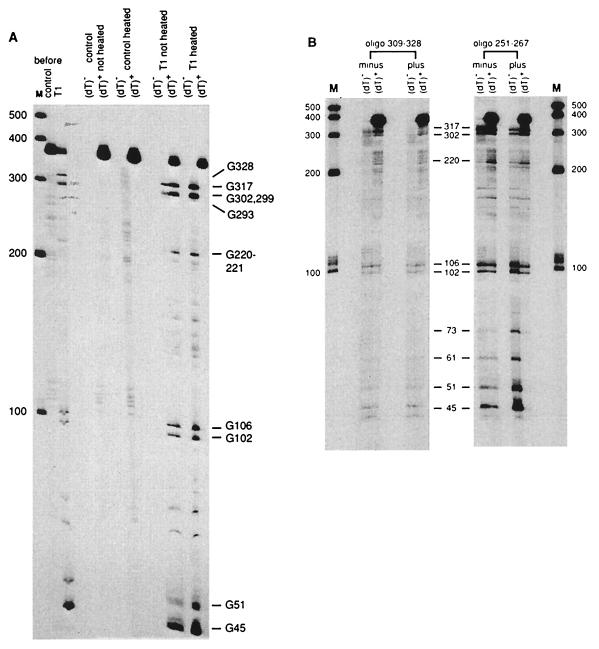
Figure 4.
Oligo(dT)-cellulose chromatography of RNase T1-cleaved MFA2 T7A38 RNA and the effect of antisense oligonucleotides. (A) T7A38 RNA (2.5 μg, prelabeled at the 5′ end) was incubated in reaction mixtures (20 μl) with no enzyme (control) or with 0.13 unit of RNase T1 for 4 min at 37°C. A 2-μl sample of each mixture was taken for gel analysis (shown at Left, control and T1, before). The reaction mixtures were then diluted with 400 μl of oligo(dT)-cellulose binding buffer (described in Materials and Methods). A 200-μl portion was heated for 3 min at 90°C, cooled rapidly, and held at 25°C for 5 min. The heated and unheated samples were then diluted and applied to oligo(dT)-cellulose columns as described in Materials and Methods. Gel analysis of the oligo(dT)-cellulose unbound (dT)− and bound (dT)+ fractions was done by using a 6% polyacrylamide gel. Ambion RNA markers are shown on the left and T1 cleavage sites on the right. (B) Labeled T7A38 RNA was annealed with and without the antisense oligonucleotides as described in Materials and Methods. The RNAs were then cleaved with 0.02 unit of RNase T1 (nucleotide 309–328 oligonucleotide) or 0.04 unit of RNase T1 (nucleotide 251–267 oligonucleotide) in 10-μl reactions. The remainder of the experiment was similar to that described in Fig. 4A for the unheated sample. T1 cleavage sites are shown in the middle and Ambion (Austin, TX) RNA markers on the left and right.