Figure 2.
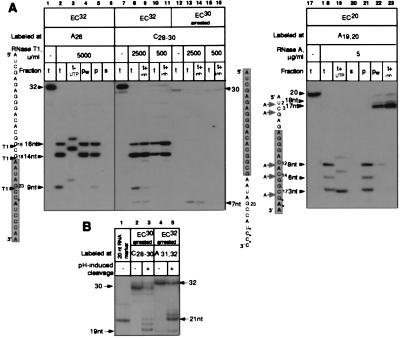
Analysis of the integrity and catalytic activity of ECs containing RNAs truncated with the RNases T1 and A. The origin of the cleavages introduced in the proximity of the RNA 3′ end. (A Left) Lanes 1–6, EC32 (lane 1) was treated with RNase T1 (lane 2, t, total). To test catalytic activity, the cleaved complex was incubated with UTP (lane 3, t+UTP). To test integrity, the cleaved complex was either washed (lane 4, pw, pellet washed) or divided into pellet and supernatant (lanes 5 and 6, p and s). Phenol was added to all the samples to inactivate the RNases. (A Center) Lanes 7–16, EC32 (lane 7) was treated with two doses of RNase T1. The cleavage was stopped either with phenol (lanes 8 and 10, t) or with 2′-GMP plus phenol (lanes 9 and 11, t+inh). Homogeneous arrested EC30 was obtained as described in Materials and Methods (lane 12) and treated with two doses of RNase T1. The cleavage was stopped either with phenol (lanes 13 and 15, t) or with 2′-GMP plus phenol (lanes 14 and 16, t+inh). (A Right) Lanes 17–23, EC20 (lane 17) was treated with RNase A and its catalytic activity and integrity were analyzed as described for A Left (lanes 18–22). Lane 23, the cleavage was stopped by Prime RNase Inhibitor plus phenol. The shaded rectangles represent the segments of the RNAs protected by RNAP in the intact ECs. (B) Homogeneous arrested EC30 and EC32 (lanes 2 and 4) were obtained as described in Materials and Methods and incubated at 37°C for 1 hr in TB adjusted to pH 9.0 by adding Tris base to stimulate the endonucleolytic activity of RNAP (27). Lane 1, 20-nt RNA used as a size marker.