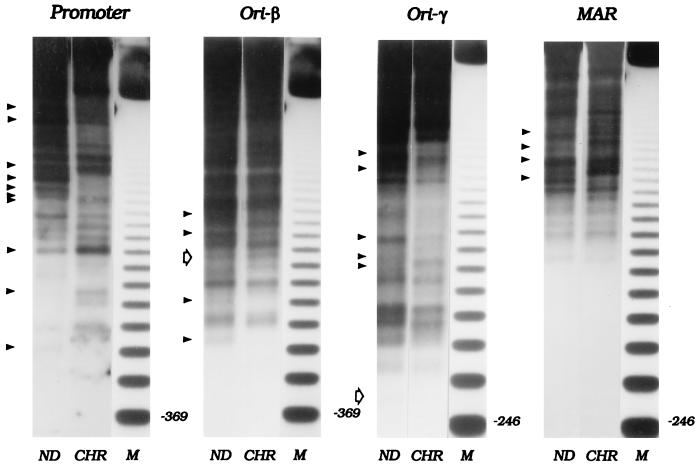
Figure 2.
Micrococcal nuclease digestion patterns of the DHFR domain in total chromatin from asynchronous CHOC 400 cells. Nuclei or naked DNA (ND) were isolated from an asynchronous culture of CHOC 400 cells and were subjected to partial micrococcal nuclease digestion as described in Materials and Methods. After digesting the DNA to completion with HindIII, the digests were separated on a 1.4% agarose gel and were transferred to Hybond N+. Digests were hybridized sequentially with probes for the indicated regions (see Materials and Methods and Fig. 1). ND, naked DNA; CHR, chromatin; M, 123-bp ladder. Solid arrowheads on the left side of each panel show the bands for which the kinetics of micrococcal nuclease digestion reproducibly differ between chromatin and naked DNA. Open arrows show the positions of cell-cycle-dependent hypersensitive sites that were detected in matrix-associated chromatin (see Fig. 5) but which are not detectable in the total chromatin samples studied in this experiment.