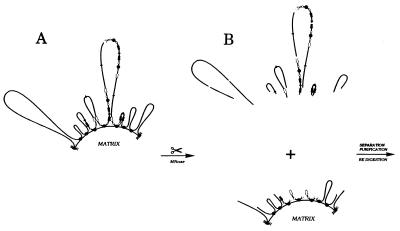
Figure 3.
Separating matrix-proximal DHFR amplicons from matrix-distal amplicons by partial micrococcal nuclease cleavage. (A) Organization of DHFR amplicons in CHOC 400 cells; all copies of the amplicon contain the intergenic MAR but only ≈15% of amplicons appear to be attached to the matrix at this site (31). Black arrows, DHFR genes; white arrows, 2BE2121 genes; filled circles, intergenic MARs; open circles, origins. (B) Micrococcal nuclease digestion was performed on intact nuclei so that subsequent manipulations to separate matrix-attached and loop fractions did not influence cut site distribution. DNA fragments from the matrix-affixed and loops fraction were purified, were digested with HindIII, and were electrophoresed on 1.4% agarose. After transfer to Hybond-N+, digests were hybridized with the probes indicated in Materials and Methods and Results and Fig. 1.