Fig. 4. FMRP target mRNAs associated with kinesin.
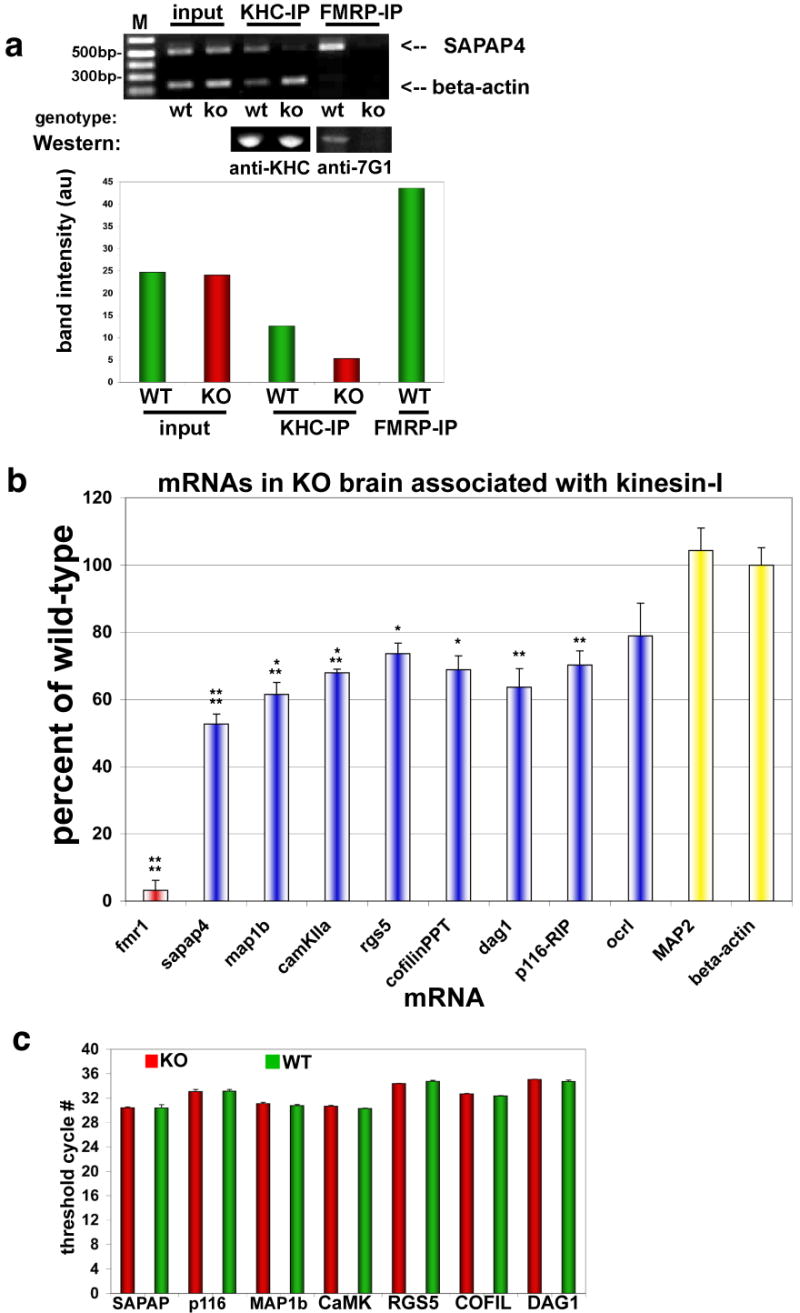
Immunoprecipitation (IP) of KHC was used to analyze associated mRNAs by PCR and determine differences between WT and FMRP KO brain.
(a) (Upper panel) Ethidium gel for semi-quantitative RT-PCR products from KHC and FMRP IP pellets using primers for beta-actin mRNA (lower bands) and SAPAP4 mRNA (upper bands). ‘Input’ is the input lysate showing starting material for both WT and KO brain. IPs of both kinesin heavy chain (KHC) and FMRP are indicated above the gel, and genotype is indicated below. FMRP IP in KO brain was used to subtract background from other bands, and was ∼4% of WT IP. (Middle panel) Western blot of protein pellets showing that the KHC IPs are identical from WT and KO brain. FMRP antibodies (anti-7G1) also reveal that FMRP is able to precipitate in WT but not KO brain (Lower panel - histogram) Quantification of mRNA bands from PCR (upper panel) by digital fluorescence analysis.
(b) Real-time Q-PCR of indicated mRNAs associated with KHC by IP, expressed as a percent of mRNA isolated from KHC in KO compared to WT brain (n=8; *p<0.05; **p<0.01; ***p<0.005; ****p<0.0001; mean±SEM).
(c) Real-time Q-PCR analysis of mRNA abundance. Histogram showing that several mRNA targets of FMRP were analyzed for total mRNA levels in both WT (green bars) and KO (red bars) brain derived from P7 mice (n=8, p≥0.4 (minimum)).
