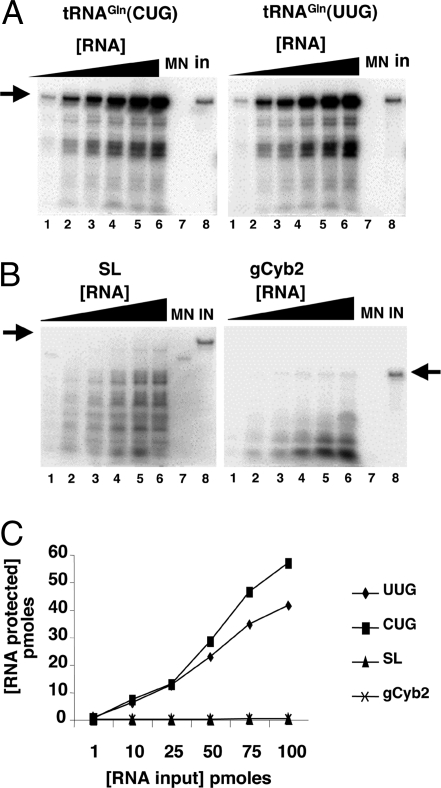
Fig. 4.
Nucleus-encoded tRNAGln is efficiently and specifically imported into isolated rat liver mitochondria in vitro. (A) Increasing concentrations of radioactively labeled ntRNACUGGln (Left) or ntRNAUUGGln transcripts (Right) were incubated with constant concentrations of Percoll-purified mitochondria as described in Materials and Methods. (B) Reactions similar to those in A but with two heterologous RNAs as specificity controls for in vitro import, where SL and gCyb2 refer to spliced leader and gRNA transcripts from trypanosomatids. These RNAs do not exist in mammalian cells. (C) A plot of the results from A and B where pmoles of input RNA protected are plotted vs. the input. MN (lanes 7), control reactions treated with micrococcal nuclease in the absence of mitochondria. These reactions serve as a control for MN digestion. IN, input lane of the original tRNA used in the import experiments and serving as a quantitation standard. The black triangle denotes increasing concentrations of a given reagent used in the assay (at 1, 10, 25, 50, 80, and 100 pmol of input, lanes 1–6 in A and B). The arrow marks the position of the full-length RNA after gel electrophoresis as described in Materials and Methods.