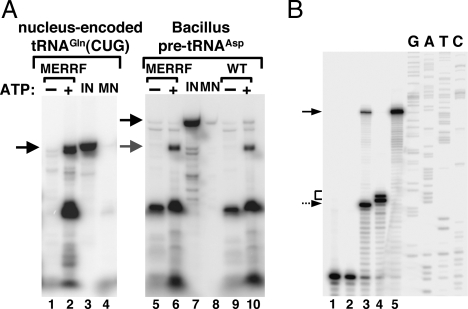
Fig. 7.
Addition of ATP rescues the ability of diseased mitochondria to import tRNAGln. (A) Radioactively labeled tRNACUGGln was incubated with mitochondria isolated from wild-type (WT) human cells, and their import behavior was compared with that isolated from cybrid cells originally generated from patients with a mitochondrial defect that causes MERRF. Mitochondria were Percoll-purified as described in Materials and Methods. Import experiments were performed in the absence (−) or presence (+) of ATP. MN, control reactions treated with micrococcal nuclease in the absence of mitochondria. These reactions serve as a control for MN digestion. IN, input lane of the original tRNA used in the import experiments and serving as a quantitation standard. Black arrows mark the position of the full-length RNA, and the gray arrow marks the position of the mature tRNA processed in the organelle following import. All import reactions were analyzed by gel electrophoresis as described in Materials and Methods. Lanes 1–4 show import reactions using a transcript corresponding to the nucleus-encoded human tRNACUGGln. Lanes 5–10 show reactions similar to those in lanes 1–4, except that pre-tRNAAsp from Bacillus was used in the import reactions to demonstrate transport of the tRNA into the matrix. (B) A reaction similar to that in lane 6 of A was analyzed by a primer extension assay to map the 5′ end of the products generated after import as described in ref. 21. Lanes 1 and 2 are negative control reactions performed in the absence of an RNA template or the absence of reverse transcriptase. Lane 3, in vitro cleavage reaction with Escherichia coli RNase P (140 pmol) incubated with the precursor tRNAAsp (40 pmol) as described in ref. 21, which yields a mature tRNA and serves as a size marker. Lane 4, result of an RNA import reaction with MERRF mitochondria and unlabeled tRNAAsp. Lane 5, full-length pre-tRNA serving as a control and size marker. G, A, T, and C denote an unrelated sequencing ladder used as size markers for the reaction. The dashed arrow corresponds to the 5′ end of pre-tRNAAsp after treatment with RNase P. The solid arrow marks the position of the pre-tRNAAsp, and the bracket marks the position of the cleavage product generated by the mitochondrial matrix localized nuclease activity found in mammalian mitochondria and described in ref. 31. This activity has been shown to digest bacterial pre-tRNAs at 2–4 nucleotides from the authentic 5′ end of the mature product (31).