FIGURE 2.
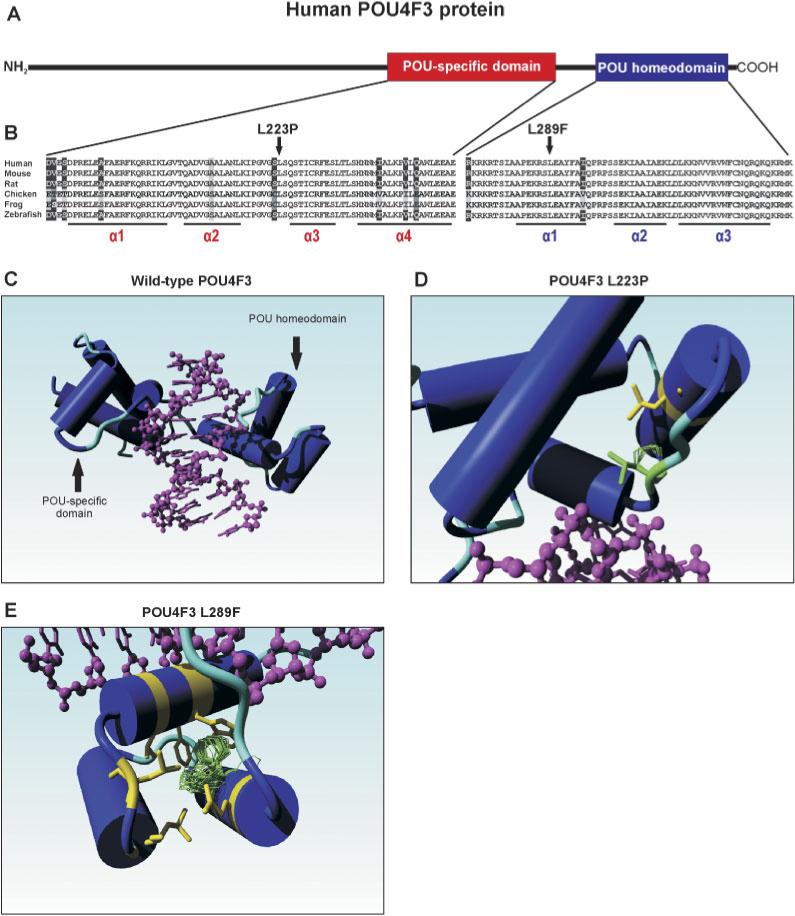
A: Schematic representation of the human POU4F3 protein.The POU-specific and the POU homeodomain are depicted in red and blue, respectively. B: Sequence comparison of the POU-specific and the POU homeodomain of several vertebrate POU4F3 proteins. Identical residues in all sequences are black on a white background, whereas amino acids that are identical in at least four species are gray on a black background. Conserved changes are black on a light gray background, whereas similar amino acids are depicted in white on a black background. The two leucine residues (L223 and L289) that were found to be substituted by a proline and phenylalanine in the two families with hearing impairment are indicated by an arrow. The positions of the various α-helices in the two DNA-binding domains are underlined. Accession numbers of the POU4F3 protein sequences: human: Q15319; mouse Q63955; rat XP 344676; chicken NP 990090; frog AAG17008; and zebrafish NP571353. C: Molecular model of the human POU4F3 protein.The part of the wild-type POU4F3 protein containing the DNA-binding domains is depicted, with the various α-helices represented as cylinders. The target DNA is indicated in purple. D: Graphic representation of the predicted effect of the p.L223P mutation. The leucine residue at position 223 is replaced by a proline (depicted in green), resulting in a clash with leucine residue 215 (in yellow) that is present in the second α-helix of the POU-specific domain. E: Graphic representation of the predicted effect of the p.L289F mutation. The leucine residue 289 is replaced by a phenylalanine. Two possible rotamer orientations of the aromatic side chain are depicted in green. In one case (upper orientation), the phenylalanine side chain clashes with the aromatic ring of a highly conserved tryptophan residue (W321) in the third α-helix of the POU homeodomain, whereas the other (less favored) orientation clashes mainly with the side-chains of two adjacent leucine residues. All residues that may somehow clash with the phenylalanine at position 289 are depicted in yellow. Images as presented in panels C–E were made using the YASARA NOVA program [Krieger et al., 2002]. [Color figure can be viewed in the online issue, which is available at http://www.interscience.wiley.com.]
