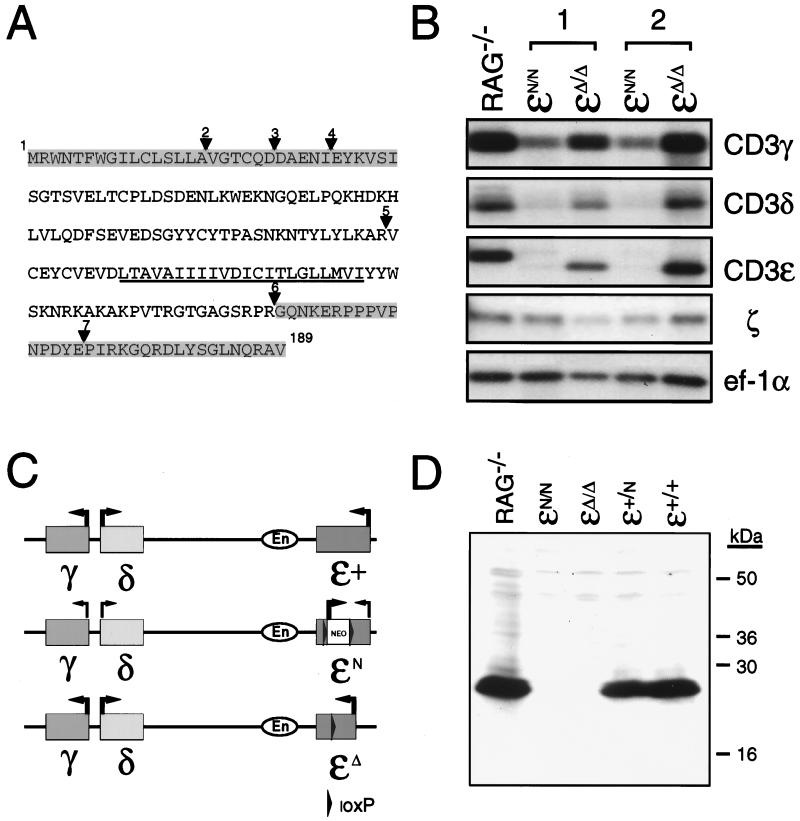
Figure 2.
(A) Predicted amino acid sequence of CD3ɛΔ. Amino acid sequence of the fully processed CD3ɛ protein is shown. The transmembrane domain is underlined. Triangles indicate the location of introns (2–7) within the CD3ɛ gene. Shaded areas represent the protein that potentially could be expressed in CD3ɛΔ/Δ mice based on the coding sequences that are retained within thymocyte transcripts from these mice. (B) Northern blot analysis of thymocyte total RNA from Rag−/− mice and from CD3ɛN/N and CD3ɛΔ/Δ mice that had been generated from two independently derived ES cell clones (1 and 2). Blots were hybridized with probes corresponding to CD3ɛ, CD3γ, CD3δ, and ζ and then were hybridized with a probe corresponding to the ubiquitously expressed ef-1α to assess the integrity and quantity of RNA in each lane. (C) Representation of the CD3-γ,-δ,-ɛ cluster on mouse chromosome 9. Location of the CD3 enhancer (En) element 3-prime of CD3ɛ is shown. Arrows indicate the direction of gene transcription. Thin lines indicate reduced gene expression. Depicted are the relative transcription levels observed from the (CD3)ɛ+, (CD3)ɛN, and (CD3)ɛΔ alleles. (D) Western blot of thymocyte protein from Rag−/−, (CD3)ɛN/N, (CD3)ɛΔ/Δ, (CD3)ɛ+/N, and (CD3)ɛ+/+ mice. Extracts from 7 × 106 thymocytes were run under reducing conditions on a 15% SDS-polyacrylamide gel, were transferred to poly(vinylidene difluoride) membrane, and were analyzed for CD3ɛ expression by using a rabbit antiserum raised against a peptide corresponding to amino acids within the C terminus of the CD3ɛ. Positions of the molecular mass standards are indicated in kilodaltons.