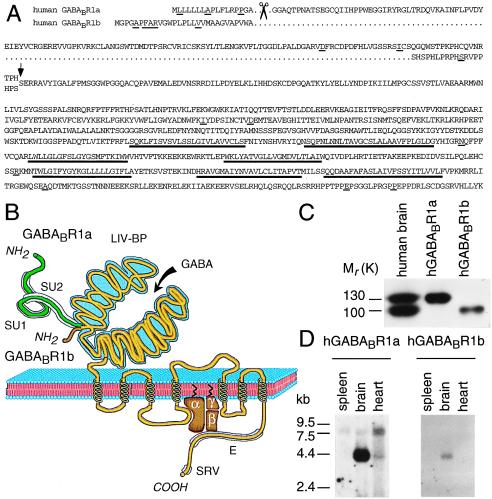
Figure 1.
Structure of hGABABRs. (A) Predicted protein sequence of hR1a and hR1b. The arrow indicates the boundary between unique N-terminal hR1a and hR1b sequences, colored in B green and brown, respectively, and the common sequence. Amino acid residues that differ between rat and human receptors are underlined. Proposed signal peptide cleavage sites are marked with scissors. Putative transmembrane domains are underlined in bold. (B) Domain structure of GABABRs. GABABRs share extended sequence similarity with mGluRs, the Ca2+-sensing receptor and a family of vomeronasal receptors (see ref. 5 and refs. therein). The mature hR1b protein differs from hR1a in that the N-terminal 147 residues are replaced by 18 different residues. The R1a-specific region mostly consists of two Sushi Repeats (SU1, SU2), ≈60 amino acid residues each, which occur in a variety of complement and adhesion proteins that engage into protein–protein interactions (42). The N-terminal domain that is common to R1a and R1b shares similarity to a guanylate cyclase intracellular domain and to bacterial periplasmic amino acid-binding proteins, such as the leucine–isoleucine–valine-binding protein (LIV-BP). Based on the three-dimensional structure of a LIV-BP, one expects this domain to fold into two lobes that constitute the GABA-binding site. The C-terminal domain is enriched in glutamic acid residues (E). At position −6 from the C terminus, GABABRs display a putative PDZ-interacting module (serine/arginine/valine, SRV) (see ref. 5 and refs. therein), which may direct the assembly of signal transduction complexes. Specificity for G protein coupling is likely provided by the second intracellular loop, as for the mGluRs. (C) [125I]CGP71872 photoaffinity labeling of membranes from rat cortex and COS-1 cells transfected with hR1a and hR1b cDNAs. Autoradiography of a SDS/PAGE (6%) is shown. (D) Northern blot analysis of hR1a and hR1b expression. Blots with 2 μg human poly(A)+ RNA per lane were hybridized to selective 32P-labeled probes.