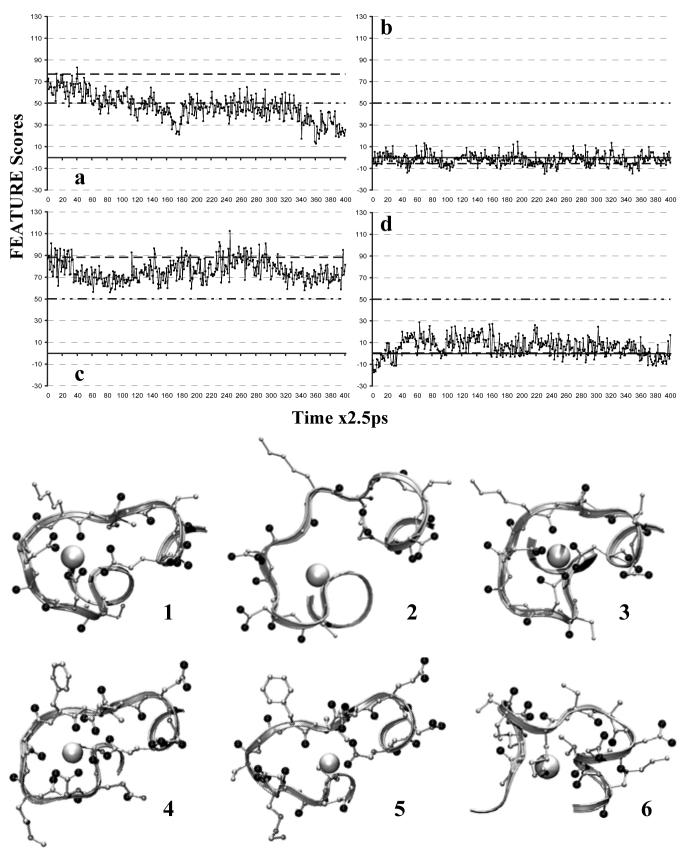
Figure 2.
FEATURE scores profiles of local grid analysis and associated structures. 1ns Simulation:  . Original FEATURE Score:
. Original FEATURE Score:  . Model Threshold:
. Model Threshold:  . a) 1B9A EF loop binding site. b) 1B9A ASP24C295, negative control. c) 1B8C EF loop binding site, positive control. d) 1B8C CD loop binding site, which was abolished by mutations. In the structures, location of the binding site is made obvious by the close association of oxygens, black balls, whether terminal ones from ASPs and GLUs or main-chains ones, near the highest-scoring points, white balls. 1) 1B9A EF loop binding site in the original static structure, which scores ∼77. 2) 1B9A EF loop binding site at 445ps, which scores ∼20. 3) 1B8C EF loop binding site at 615ps, which scores ∼112. 4) 1B9A CD loop binding site at 184ps, which scores ∼52. 5) 1B8C CD loop binding site, abolished by the mutations, at 187.5ps, which scores ∼13.5. 6) 1B9A negative control centered on ASP24C295 at 805ps, which scores ∼13.5.
. a) 1B9A EF loop binding site. b) 1B9A ASP24C295, negative control. c) 1B8C EF loop binding site, positive control. d) 1B8C CD loop binding site, which was abolished by mutations. In the structures, location of the binding site is made obvious by the close association of oxygens, black balls, whether terminal ones from ASPs and GLUs or main-chains ones, near the highest-scoring points, white balls. 1) 1B9A EF loop binding site in the original static structure, which scores ∼77. 2) 1B9A EF loop binding site at 445ps, which scores ∼20. 3) 1B8C EF loop binding site at 615ps, which scores ∼112. 4) 1B9A CD loop binding site at 184ps, which scores ∼52. 5) 1B8C CD loop binding site, abolished by the mutations, at 187.5ps, which scores ∼13.5. 6) 1B9A negative control centered on ASP24C295 at 805ps, which scores ∼13.5.