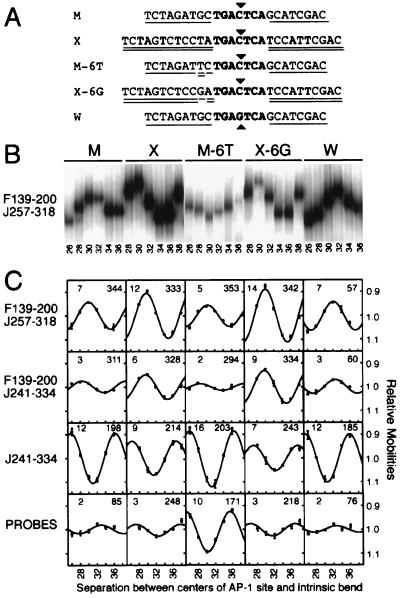
Figure 2.
Effects of sequences flanking the AP-1 site on DNA bending by the bZIP domains of Fos and Jun. (A) Sequences of AP-1 sites analyzed. Flanking sequences shared with the M site are single underlined whereas flanking sequences shared with the X site are double underlined. The core AP-1 recognition element is shown in boldface. A central C:G base pair is indicated by a triangle above the sequence whereas a central G:C base pair is indicated by a triangle below the sequence. (B) Phasing analysis of Fos–Jun bZIP domain heterodimers bound to different AP-1 sites. Fos139–200 and Jun257–318 were incubated with phasing analysis probes containing the sites shown above, and the complexes were analyzed by PAGE. The complexes were run on the same gel and the lanes were rearranged for a more concise figure. (C) The average mobilities of complexes formed by Fos139–200—Jun257–318, Fos139–200—Jun241–334, and Jun241–334 on different binding sites were normalized for differences in probe mobilities (shown at the bottom of the figure) and were plotted as a function of the separation between the centers of the AP-1 site and the intrinsic bend. The DNA bend angle and direction calculated based on the mobility variation are indicated in the upper left and upper right hand corners of each plot. Standard deviations of the mobilities in multiple independent experiments are plotted for each complex. Multivariate analysis of the variance in DNA bending for all of the complexes indicated that the differences in DNA bending were highly significant (probability of identity < 0.001) between all pairs of sites.