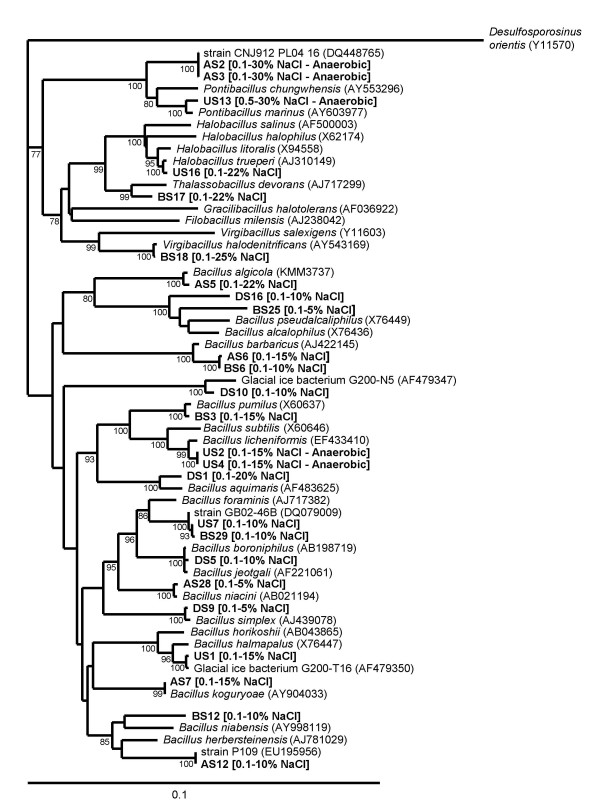
Figure 2.
Phylogenetic tree based on 16S rRNA sequences of isolates from l'Atalante (AS), Bannock (BS), Discovery (DS) and Urania (US) brine-lake sediments. The tree was constructed based on an alignment of approximately 1340 base pairs from the isolates and their closest relatives. Uncharacterized strains were included in the tree when there was no closely related named species. For the named species, the sequence from the type strain was used. Accession numbers of reference strains are given in round brackets. The scale bar corresponds to 0.1 substitutions per nucleotide position. Figures (%) at branching points represent significance of branching order by bootstrap analysis (1000 replicates, bootstrap values above 75% are shown). For the strains isolated from the deep-sea brine-lake sediments, the NaCl range for growth and the ability to grow under anaerobic conditions are indicated in square brackets. Note that while strain US13 did not grow fermentatively, it grew using dimethylsulfoxide or trimethylamine N-oxide as electron acceptor.