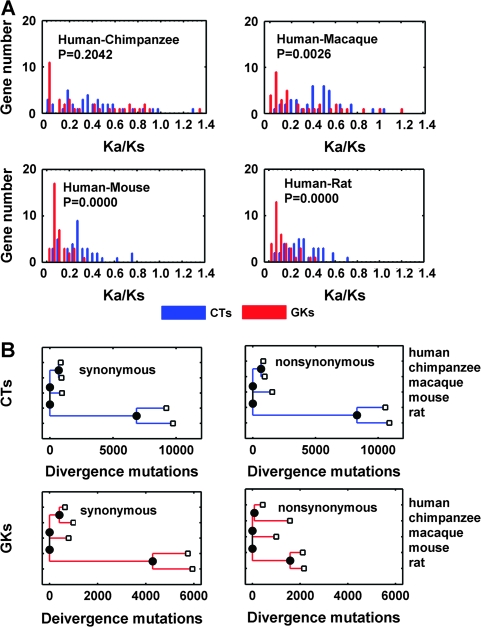
FIG. 2.—
Evolutionary rate comparison of CTs and GKs. (A) Distribution of Ka/Ks in pairwise comparisons between CTs and GKs within 4 lineages including human–chimpanzee (top left), human–macaca (top right), human–mouse (bottom left), and human–rat (bottom right). Bin size 0.05 was used for distribution computation. For this analysis—which compares the different evolutionary rates of human CTs and GKs using a variety of species comparators, as distinct from comparing gene evolutionary rates in multiple species—P values were calculated by ranked 2-sample Mann–Whitney U test using MATLAB Statistical toolbox function rank sum. (B) Lineage-specific comparison of evolutionary rate of CTs and GKs using supergene concatenation. Blue bars, CTs; red bars, GKs.