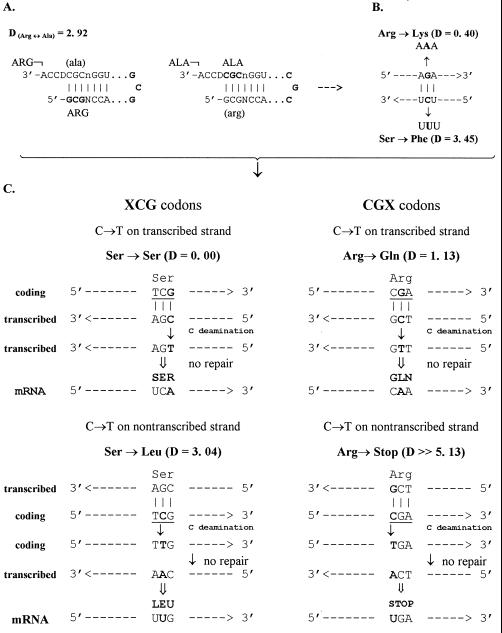
Figure 3.
(A) Hypothetical pair of tRNAArg and tRNAAla with complementary anticodons originated from the opposite strands of the same acceptor. This pair looks extremely disadvantageous because incorrect aminoacylation, quite likely for identical acceptors, had to involve very dissimilar amino acids, thus leading to large pleiotropical perturbations of all early proteins. (B) The above second base-associated disadvantage, inherent in the situation (A), appears as an important constraint on the primordial double-strand coding. Shown is an example of quite conservative mutation (Arg → Lys) caused by a G → A transition at the codon second position in one strand, inevitably accompanied by a detrimental mutation (Ser → Phe) caused by symmetric C → T transition in the complementary strand. Analysis of the code (Table 1) shows that most of all such pairs of complementary base substitutions result in either one or even both detrimental mutations. (C) Two examples showing that of the two DNA strands, the coding nontranscribed one is, on average, more vulnerable regarding protein function when the C → T transition originates at the hypermutable CpG dinucleotides within TCG and CGA codons, respectively. Nonconservative missense mutation Ser → Leu in TCG codon on the nontranscribed (coding) strand is complemented by a silent mutation on the opposite transcribed (noncoding) strand; similarly, detrimental Arg → Stop mutation in CGA codon on the nontranscribed strand is mirrored by an evidently more conservative Arg → Gln missense mutation on the transcribed strand. All other CpG-containing codons (three XCG and three CGX) exhibit the same strand asymmetry. Its application to the comparison of different tumor-attributed versus evolutionary mutation databases will be published separately.