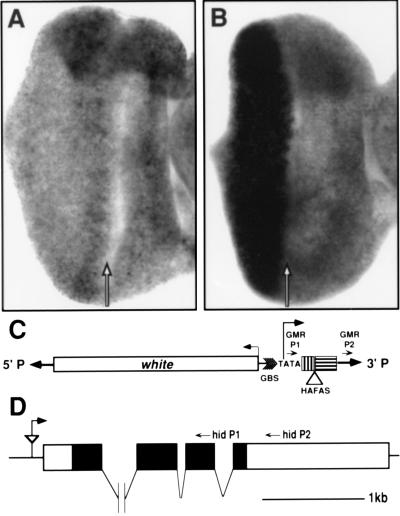
Figure 3.
Misexpression of the hid gene by the GMR-HAFAS110 insertion. The hid expression pattern in eye-antennal discs from wild type (A) and GMR-HAFAS110/GMR-HAFAS110 (B) larvae are shown. The hid transcript is expressed uniformly in the wild-type eye imaginal disc (A) and at high levels in and posterior (left) to the morphogenetic furrow (arrow) in GMR-HAFAS110 eye imaginal discs (B). (C) Map of the GMR-HAFAS construct. The expression vector pGMR contains the white marker gene (white), a multimer of glass binding sites (GBS), TATA sequences from the hsp70 promoter (TATA), approximately 200 bases of 5′ untranslated region (box marked with vertical bars), a polylinker into which the HAFAS construct was cloned (HAFAS is not drawn to scale), and the hsp 70 3′ untranslated region (box marked with horizontal bars). The direction of transcription from the hsp70 promoter is indicated. The P element ends are indicated as boldface arrows. The location of the P element primers used to detect chimeric transcripts are indicated with small arrowheads (GMR P1 and GMR P2). (D) Map of the genomic region at the site of insertion of the GMR-HAFAS110 P element. GMR-HAFAS110 is inserted 131 base pairs 5′ to the longest hid cDNA (described in ref. 26). The P element is indicated by the triangle and is oriented such that transcription from the hsp70 TATA box reads through the P element 3′ end and into the 5′ end of the hid transcription unit as indicated by the arrow. The locations of primers used for cDNA synthesis (hid P2) and PCR (hid P1) are indicated on the map. The introns are not drawn to scale.