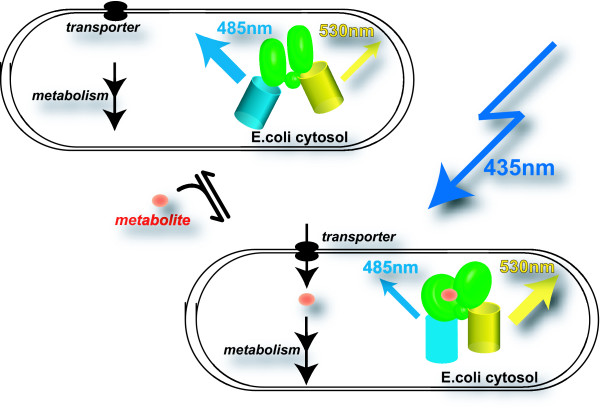
Figure 1.
In vivo analysis of metabolite accumulation in live bacterial cells using fluorescence resonance energy transfer sensors. Gram-negative bacterial cells are shown surrounded by two membranes expressing the fluorescence resonance energy transfer sensor, which consists of an episomally encoded recognition element (green central domain composed of two lobes that both contribute to ligand binding, connected by a hinge region), here a periplasmic binding protein for arabinose or maltose, sandwiched by an N-terminal cyan variant of green fluorescent protein (enhanced cyan fluorescent protein) and a C-terminal yellow variant (enhanced yellow fluorescent protein). The sensor contains no targeting information and will thus be present in the cytosol of Escherichia coli. Excitation of enhanced cyan fluorescent protein with 435 nm light results in emission from enhanced cyan fluorescent protein (peak emission at ~485 nm) and fluorescence resonance energy transfer-derived enhanced yellow fluorescent protein emission (peak emission at ~528 nm). The important characteristics of the sensor are the binding constant and detection range. The cell in the top panel is cultivated in the absence of the ligand; therefore, endogenous ligand concentrations are low (all sensors are in the unbound state, with low fluorescence resonance energy transfer). The cell in the bottom panel is cultivated in the presence of the ligand; therefore, endogenous ligand concentrations are high (all sensors are in the bound state, with high fluorescence resonance energy transfer). The steady-state levels and accumulation rates in the cytosol will be determined predominantly by the external ligand levels as well as the properties and abundance of transporters and the downstream metabolic enzymes.