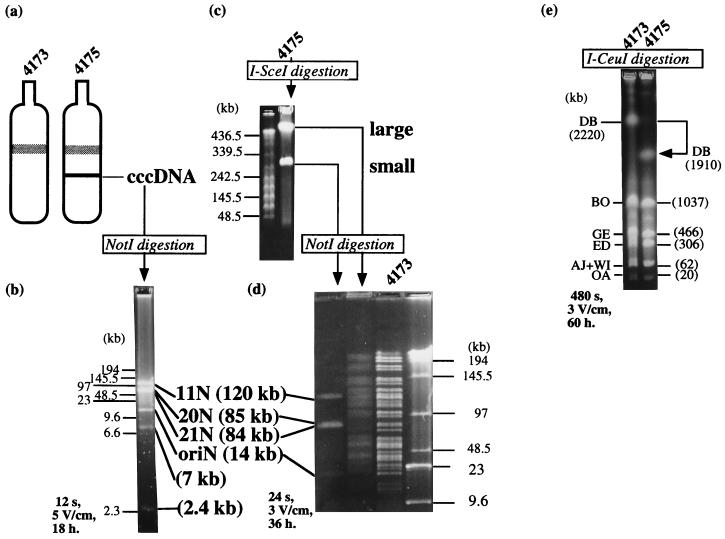
Figure 3.
Isolation and characterization of the 310-kb subgenome. (a) A total DNA lysate gently prepared from 10-ml cultures of BEST4173 and BEST4175 were centrifuged in a CsCl-ethidium bromide gradient at 140,000 × g for 18 hr in the vertical rotor using Beckman L-80 ultracentrifuge. Only the BEST4175 sample gave an extra band that was subsequently recovered, desalted and destained by extensive dialysis against TE solution (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA). (b) The cccDNA sample was digested with NotI and resolved by pulsed-field gel electrophoresis. The cccDNA gave rise to six NotI segments that are predicted in Fig. 2. (c) I-SceI digestion of BEST4175 DNA gave two linear DNA fragments in pulsed-field gel electrophoresis. The large and small DNAs were isolated and subjected to NotI digestion. Concatemeric lambda DNA (48.5 × n kb) was used as size markers. (d) The small I-SceI fragment gave NotI fragments identical to those of the cccDNA. The 7- and 2.4-kb NotI fragments were verified for their presence in a separate experiment (data not shown). A NotI digest of the large I-SceI fragment was resolved in pulsed-field gel electrophoresis and found to be composed of the genomic NotI fragments lacking the subgenome fragments, as shown by the comparison with the NotI fragments of BEST4173 DNA in the next lane. Concatemeric lambda DNA (48.5 × n kb) and HindIII digested lambda DNA are size markers. (e) Total DNAs from BEST4173 and BEST4175 were digested with I-CeuI. The DB fragment derived from the I-CeuI sites in the rrnD and rrnB operons shown in Fig. 1. In BEST4175 only the 2,220-kb DB fragment was shortened to the 1,910-kb band, which verifies that the main genome carried a deletion of a DNA segment with the size equivalent to the subgenome. The subgenome remained as circular DNA and did not enter the gel. Specific running conditions for pulsed-field gel electrophoresis, pulse time, voltages, and running time are shown in each figure.