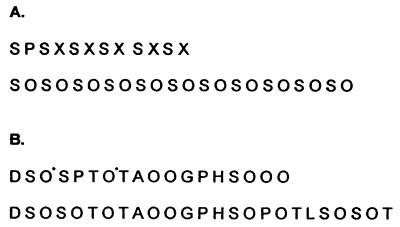Figure 5.
Polypeptide sequences of (Ser-Hyp)32-EGFP and (GAGP)3-EGFP before and after deglycosylation. (A) N-terminal amino acid sequence of the glycoprotein (Ser-Hyp)32-EGFP. We obtained partial sequence of both the glycoprotein (Upper) and its polypeptide after deglycosylation (Lower). X denotes blank cycles that correspond to glycosylated Hyp; glycoamino acids tend to produce blank cycles during Edman degradation, an exception being arabinosyl Hyp (17). (B) Polypeptide sequence of glycosylated (GAGP)3-EGFP (Upper) and deglycosylated (GAGP)3-EGFP (Lower). Residues marked with an asterisk denote low molar yields of Hyp and likely sites of arabinogalactan polysaccharide attachment in glycosylated (GAGP)3-EGFP. For example, yields were 480 pM Asp in the first cycle, 331 pM Ser in the second, 194 pM Hyp in the third, and 508 pM Ser in the fourth.