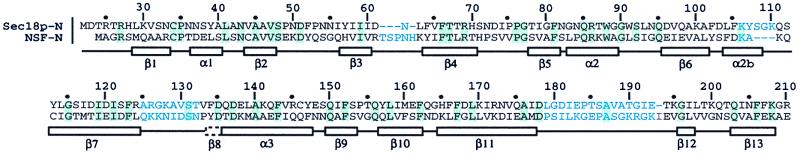
Figure 1.
Sequence alignment of S. cerevisiae Sec18p-N and hamster NSF-N based on a visual comparison of the two structures. Sequence alignments performed before knowledge of the Sec18p-N structure introduced gaps between strands β2 and β3 in subdomain NA. Such gaps were hypothesized to be inconsistent with a double-psi β-barrel (33). In fact, the only gap in the proper alignment of NA occurs in the loop between strands β3 and β4, which is elongated in NSF vs. Sec18p by an additional four residues. Insertions in this loop are proposed to be tolerated in the double-psi β-barrel (33). The Sec18p-N sequence is labeled every 10 residues according to the full-length Sec18p numbering. Secondary structural elements are boxed below the sequences. The box with a dashed border indicates a short strand (β8) present in NSF-N but not observed in Sec18p-N. Sequence identities are highlighted in light green. Sequences shown in light blue indicate regions with structures that diverge significantly between NSF-N and Sec18p-N, or that were not built in one or the other structural models.