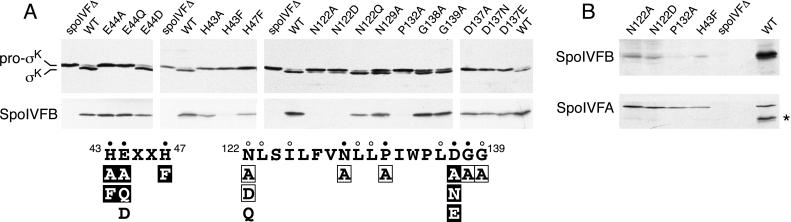
Figure 2.
Pro-σK processing in SpoIVFB mutants. (A) Immunoblots of whole-cell extracts from sporulating cells. All strains analyzed contained a deletion of spoIVF at its normal locus (spoIVFΔ) and a copy of the spoIVF operon at the amyE locus containing either wild-type spoIVFB (WT) or the indicated spoIVFB point mutation. (Upper) Extracts from hour 5 sporulating cells were analyzed by using antibodies that recognize both pro-σK and σK. (Lower) Extracts from hour 3 sporulating cells were analyzed by using antibodies that recognize the C terminus of SpoIVFB (a region that does not contain the two motifs). The results of the analysis are shown schematically below the immunoblots. The amino acid sequences of the two conserved motifs in SpoIVFB are shown in black. Amino acid positions are indicated. Closed and open dots indicate amino acids that are virtually invariant and highly conserved, respectively. Amino acid substitutions that completely abolished pro-σK processing are in black boxes. Mutations that reduced processing efficiency are in black and are surrounded by a box, and those that had no detectable effect on processing are in black. (B) An immunoblot of the four point mutants that had no detectable SpoIVFB protein in A. Twenty-five percent more extract (from hour 3 sporulating cells) was used in this experiment than in those in A. The immunoblot was analyzed by using anti-SpoIVFB antibodies (Upper) and was overexposed to detect the low level of SpoIVFB mutant protein. The immunoblot was reprobed with antibodies that recognize SpoIVFA to control for loading (Lower). The asterisk (*) indicates the position of residual SpoIVFB signal on the reprobed blot.