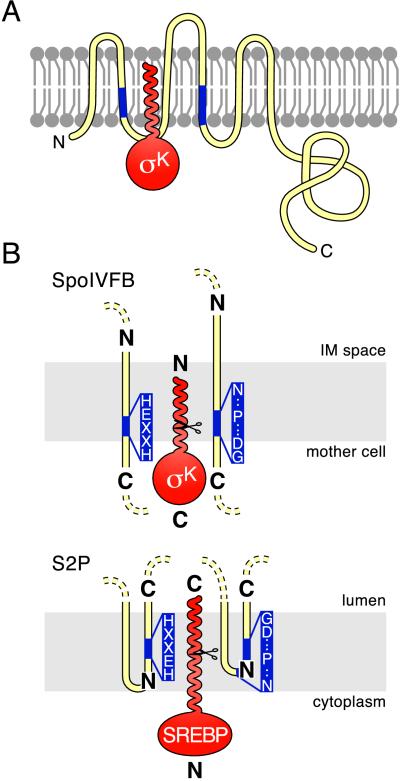
Figure 4.
Topological models for SpoIVFB, S2P and their substrates. Membranes are shown schematically in gray. Pro-σK and SREBP are in red. SpoIVFB and S2P are in yellow with their conserved motifs in blue (the invariant residues in the two motifs are shown in B). N and C termini are indicated. (A) Proposed insertion of the pro domain of pro-σK into the membrane. Pro-σK is shown with SpoIVFB. The membrane topology of SpoIVFB is based on the work of Green and Cutting (37). The N-terminal pro domain of pro-σK is drawn as a helix based on its predicted secondary structure (40). The mother-cell cytoplasm is below the membrane, and the space between the mother-cell and forespore membranes (the intermembrane space) is above it. (B) Proposed orientation of the catalytic centers in SpoIVFB and S2P. Relevant membrane segments of SpoIVFB and S2P are shown relative to the mother cell or cytoplasmic face of the membrane and to the intermembrane space (IM) or the lumen. Scissors indicate cleavage sites in pro-σK and SREBP. To accommodate the opposite orientation of the transcription factors, the catalytic centers in the two processing enzymes are shown in opposite orientations relative to the compartment into which the mature transcription factors are released (see text).